4PPD
 
 | | PduA K26A, crystal form 2 | | Descriptor: | GLYCEROL, Propanediol utilization protein PduA, SULFATE ION | | Authors: | McNamara, D.E, Sawaya, M.R, Bobik, T.A, Yeates, T.O. | | Deposit date: | 2014-02-26 | | Release date: | 2014-05-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Alanine scanning mutagenesis identifies an asparagine-arginine-lysine triad essential to assembly of the shell of the pdu microcompartment.
J.Mol.Biol., 426, 2014
|
|
2CG7
 
 | | SECOND AND THIRD FIBRONECTIN TYPE I MODULE PAIR (CRYSTAL FORM II). | | Descriptor: | FIBRONECTIN | | Authors: | Rudino-Pinera, E, Ravelli, R.B.G, Sheldrick, G.M, Nanao, M.H, Werner, J.M, Schwarz-Linek, U, Potts, J.R, Garman, E.F. | | Deposit date: | 2006-02-27 | | Release date: | 2007-02-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The Solution and Crystal Structures of a Module Pair from the Staphylococcus Aureus-Binding Site of Human Fibronectin-A Tale with a Twist.
J.Mol.Biol., 368, 2007
|
|
8EMV
 
 | | Phospholipase C beta 3 (PLCb3) in solution | | Descriptor: | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase beta-3, CALCIUM ION | | Authors: | Falzone, M.E, MacKinnon, R. | | Deposit date: | 2022-09-28 | | Release date: | 2023-05-24 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | G beta gamma activates PIP2 hydrolysis by recruiting and orienting PLC beta on the membrane surface.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
1J5O
 
 | | CRYSTAL STRUCTURE OF MET184ILE MUTANT OF HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH DOUBLE STRANDED DNA TEMPLATE-PRIMER | | Descriptor: | 5'-D(*AP*TP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*C)-3', 5'-D(*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*GP*CP*GP*CP*CP*A)-3', ANTIBODY (HEAVY CHAIN), ... | | Authors: | Sarafianos, S.G, Das, K, Arnold, E. | | Deposit date: | 2002-05-24 | | Release date: | 2002-06-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Lamivudine (3TC) resistance in HIV-1 reverse transcriptase involves steric hindrance with beta-branched amino acids.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
6EIA
 
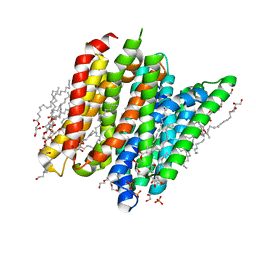 | | PepTSt in complex with HEPES (100 mM) | | Descriptor: | (2R)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, (2S)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Martinez Molledo, M, Quistgaard, E.M, Loew, C. | | Deposit date: | 2017-09-18 | | Release date: | 2018-02-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Multispecific Substrate Recognition in a Proton-Dependent Oligopeptide Transporter.
Structure, 26, 2018
|
|
1QJE
 
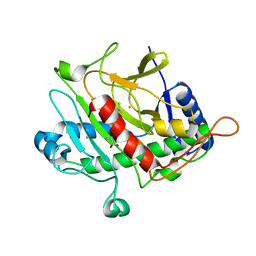 | | Isopenicillin N synthase from Aspergillus nidulans (IP1 - Fe complex) | | Descriptor: | FE (II) ION, ISOPENICILLIN N, ISOPENICILLIN N SYNTHASE, ... | | Authors: | Burzlaff, N.I, Clifton, I.J, Rutledge, P.J, Roach, P.L, Adlington, R.M, Baldwin, J.E. | | Deposit date: | 1999-06-23 | | Release date: | 2000-06-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The Reaction Cycle of Isopenicillin N Synthase Observed by X-Ray Diffraction
Nature, 401, 1999
|
|
2IJH
 
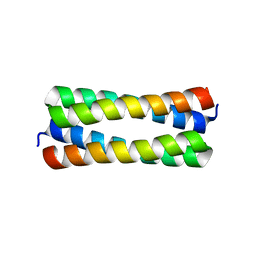 | | Crystal structure analysis of ColE1 ROM mutant F14W | | Descriptor: | Regulatory protein rop | | Authors: | Ladner, J.E. | | Deposit date: | 2006-09-29 | | Release date: | 2007-10-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | New crystal structures of ColE1 Rom and variants resulting from mutation of a surface exposed residue: Implications for RNA-recognition.
Proteins, 72, 2008
|
|
6EJC
 
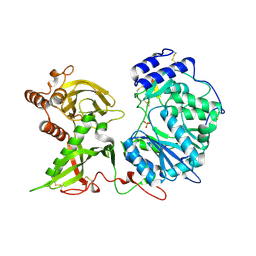 | |
2CHZ
 
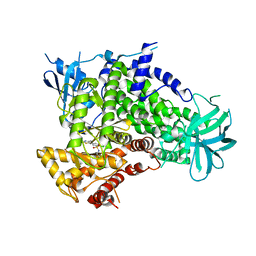 | | A pharmacological map of the PI3-K family defines a role for p110alpha in signaling: The structure of complex of phosphoinositide 3-kinase gamma with inhibitor PIK-93 | | Descriptor: | N-(5-(4-CHLORO-3-(2-HYDROXY-ETHYLSULFAMOYL)- PHENYLTHIAZOLE-2-YL)-ACETAMIDE, PHOSPHATIDYLINOSITOL-4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT GAMMA ISOFORM | | Authors: | Knight, Z.A, Gonzalez, B, Feldman, M.E, Zunder, E.R, Goldenberg, D.D, Williams, O, Loewith, R, Stokoe, D, Balla, A, Toth, B, Balla, T, Weiss, W.A, Williams, R.L, Shokat, K.M. | | Deposit date: | 2006-03-16 | | Release date: | 2006-05-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A Pharmacological Map of the Pi3-K Family Defines a Role for P110Alpha in Signaling
Cell(Cambridge,Mass.), 125, 2006
|
|
8EMS
 
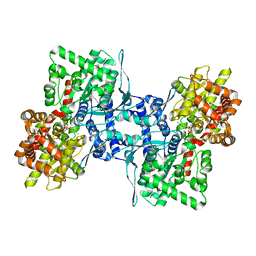 | | Cryo-EM structure of human liver glycogen phosphorylase | | Descriptor: | Glycogen phosphorylase, liver form, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Su, C, Lyu, M, Zhang, Z, Yu, E.W. | | Deposit date: | 2022-09-28 | | Release date: | 2023-05-10 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | High-resolution structural-omics of human liver enzymes.
Cell Rep, 42, 2023
|
|
2ISA
 
 | | Crystal Structure of Vibrio salmonicida catalase | | Descriptor: | CHLORIDE ION, Catalase, GLYCEROL, ... | | Authors: | Riise, E.K, Lorentzen, M.S, Helland, R, Smalas, A.O, Leiros, H.K.S, Willassen, N.P. | | Deposit date: | 2006-10-17 | | Release date: | 2007-01-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | The first structure of a cold-active catalase from Vibrio salmonicida at 1.96A reveals structural aspects of cold adaptation
ACTA CRYSTALLOGR.,SECT.D, 63, 2007
|
|
6EM1
 
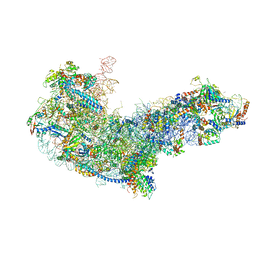 | | State C (Nsa1-TAP Flag-Ytm1) - Visualizing the assembly pathway of nucleolar pre-60S ribosomes | | Descriptor: | 25S ribosomal RNA, 5.8S ribosomal RNA, 60S ribosomal protein L13-A, ... | | Authors: | Kater, L, Cheng, J, Barrio-Garcia, C, Hurt, E, Beckmann, R. | | Deposit date: | 2017-10-01 | | Release date: | 2017-12-27 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Visualizing the Assembly Pathway of Nucleolar Pre-60S Ribosomes.
Cell, 171, 2017
|
|
8OKP
 
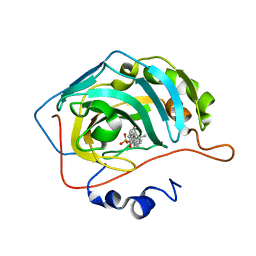 | | Carbonic Anhydrase IX like mutant in Complex with Steriod_Sulphamoyl inhibitor AKI_33 | | Descriptor: | Carbonic anhydrase 2, ZINC ION, [(3~{R},5~{R},8~{R},9~{S},10~{S},13~{S},14~{S},15~{R})-10,13,15-trimethyl-17-oxidanylidene-1,2,3,4,5,6,7,8,9,11,12,14,15,16-tetradecahydrocyclopenta[a]phenanthren-3-yl] sulfamate | | Authors: | Brynda, J, Rezacova, P.M, Kudova, E. | | Deposit date: | 2023-03-28 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Carbonic Anhydrase II in Complex with Steriod_Sulphamoyl
To Be Published
|
|
6EF2
 
 | | Yeast 26S proteasome bound to ubiquitinated substrate (5T motor state) | | Descriptor: | 26S proteasome regulatory subunit 4 homolog, 26S proteasome regulatory subunit 6A, 26S proteasome regulatory subunit 6B homolog, ... | | Authors: | de la Pena, A.H, Goodall, E.A, Gates, S.N, Lander, G.C, Martin, A. | | Deposit date: | 2018-08-15 | | Release date: | 2018-10-17 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.27 Å) | | Cite: | Substrate-engaged 26Sproteasome structures reveal mechanisms for ATP-hydrolysis-driven translocation.
Science, 362, 2018
|
|
7NBV
 
 | | Structure of 2A protein from Theilers murine encephalomyelitis virus (TMEV) | | Descriptor: | BROMIDE ION, Capsid protein VP0 | | Authors: | Hill, C.H, Cook, G.M, Napthine, S, Kibe, A, Brown, K, Caliskan, N, Firth, A.E, Graham, S.C, Brierley, I. | | Deposit date: | 2021-01-28 | | Release date: | 2021-12-08 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Investigating molecular mechanisms of 2A-stimulated ribosomal pausing and frameshifting in Theilovirus.
Nucleic Acids Res., 49, 2021
|
|
2CC1
 
 | |
8OKT
 
 | | Carbonic Anhydrase IX like mutant in Complex with Steriod_Sulphamoyl inhibitor VK42 | | Descriptor: | Carbonic anhydrase 2, ZINC ION, [(8~{R},9~{S},13~{S},14~{S})-13-methyl-17-oxidanylidene-7,8,9,11,12,14,15,16-octahydro-6~{H}-cyclopenta[a]phenanthren-3-yl] sulfamate | | Authors: | Brynda, J, Rezacova, P.M, Kudova, E. | | Deposit date: | 2023-03-29 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Carbonic Anhydrase II in Complex with Steriod_Sulphamoyl
To Be Published
|
|
8OMN
 
 | | Carbonic Anhydrase II in Complex with Steriod_Sulphamoyl VK4 | | Descriptor: | Carbonic anhydrase 2, ZINC ION, [(3~{R},5~{R},8~{R},9~{S},10~{S},13~{S},14~{S},17~{S})-17-ethanoyl-10,13-dimethyl-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1~{H}-cyclopenta[a]phenanthren-3-yl] sulfamate | | Authors: | Brynda, J, Rezacova, P.M, Kudova, E. | | Deposit date: | 2023-03-31 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Carbonic Anhydrase II in Complex with Steriod_Sulphamoyl
To Be Published
|
|
6II6
 
 | | Crystal structure of the Makes Caterpillars Floppy (MCF)-Like effector of Vibrio vulnificus MO6-24/O in complex with a human ADP-ribosylation factor 3 (ARF3) | | Descriptor: | ADP-ribosylation factor 3, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Lee, Y, Kim, B.S, Choi, S, Lee, E.Y, Park, S, Hwang, J, Kwon, Y, Hyun, J, Lee, C, Eom, S.H, Kim, M.H. | | Deposit date: | 2018-10-03 | | Release date: | 2019-08-07 | | Last modified: | 2020-02-19 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Makes caterpillars floppy-like effector-containing MARTX toxins require host ADP-ribosylation factor (ARF) proteins for systemic pathogenicity.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6EBG
 
 | | Ohr (Organic Hydroperoxide Resistance protein) mutant - C60S interacting with dihydrolipoamide | | Descriptor: | (6S)-6,8-disulfanyloctanamide, Organic hydroperoxide resistance protein | | Authors: | Domingos, R.M, Teixeira, R.D, Alegria, T.G.P, Vieira, P.S, Murakami, M.T, Netto, L.E.S. | | Deposit date: | 2018-08-06 | | Release date: | 2020-02-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Substrate and product-assisted catalysis: molecular aspects behind structural switches along Organic Hydroperoxide Resistance Protein catalytic cycle
Acs Catalysis, 2020
|
|
4ZVT
 
 | |
3HE5
 
 | |
2CO4
 
 | | Salmonella enterica SafA pilin in complex with a 19-residue SafA Nte peptide | | Descriptor: | PUTATIVE OUTER MEMBRANE PROTEIN | | Authors: | Remaut, H, Rose, R.J, Hannan, T.J, Hultgren, S.J, Radford, S.E, Ashcroft, A.E, Waksman, G. | | Deposit date: | 2006-05-25 | | Release date: | 2006-06-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Donor-Strand Exchange in Chaperone-Assisted Pilus Assembly Proceeds Through a Concerted Beta-Strand Displacement Mechanism
Mol.Cell, 22, 2006
|
|
8OKO
 
 | | Carbonic Anhydrase IX like mutant in Complex with Steriod_Sulphamoyl inhibitor AKI_2 | | Descriptor: | Carbonic anhydrase 2, ZINC ION, [(3~{S},8~{S},9~{S},10~{R},13~{S},14~{S})-10,13-dimethyl-2,3,4,7,8,9,11,12,14,15,16,17-dodecahydro-1~{H}-cyclopenta[a]phenanthren-3-yl] sulfamate | | Authors: | Brynda, J, Rezacova, P.M, Kudova, E. | | Deposit date: | 2023-03-28 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Carbonic Anhydrase II in Complex with Steriod_Sulphamoyl
To Be Published
|
|
2CFV
 
 | | Crystal structure of human protein tyrosine phosphatase receptor type J | | Descriptor: | CHLORIDE ION, HUMAN PROTEIN TYROSINE PHOSPHATASE RECEPTOR TYPE J, NICKEL (II) ION | | Authors: | Debreczeni, J.E, Barr, A.J, Eswaran, J, Ugochukwu, E, Sundstrom, M, Weigelt, J, Arrowsmith, C, Edwards, A, Knapp, S. | | Deposit date: | 2006-02-23 | | Release date: | 2006-03-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Large-Scale Structural Analysis of the Classical Human Protein Tyrosine Phosphatome.
Cell(Cambridge,Mass.), 136, 2009
|
|
