7UPL
 
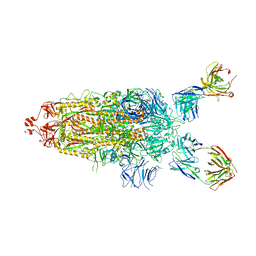 | | SARS-Cov2 Omicron varient S protein structure in complex with neutralizing monoclonal antibody 002-S21F2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Patel, A, Ortlund, E. | | Deposit date: | 2022-04-15 | | Release date: | 2022-08-10 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural insights for neutralization of Omicron variants BA.1, BA.2, BA.4, and BA.5 by a broadly neutralizing SARS-CoV-2 antibody.
Sci Adv, 8, 2022
|
|
7X1M
 
 | | The complex structure of Omicron BA.1 RBD with BD604, S309,and S304 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, BD-604 Fab heavy chain, BD-604 Fab light chain, ... | | Authors: | Huang, M, Xie, Y.F, Qi, J.X. | | Deposit date: | 2022-02-24 | | Release date: | 2022-07-06 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | Atlas of currently available human neutralizing antibodies against SARS-CoV-2 and escape by Omicron sub-variants BA.1/BA.1.1/BA.2/BA.3.
Immunity, 55, 2022
|
|
7RG8
 
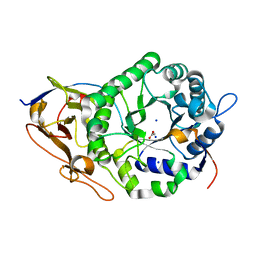 | | Crystal Structure of a Stable Heparanase Mutant | | Descriptor: | ACETATE ION, Heparanase 50 kDa subunit, Heparanase 8 kDa subunit, ... | | Authors: | Whitefield, C, Hong, N.S, Jackson, C.J. | | Deposit date: | 2021-07-14 | | Release date: | 2022-03-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Computational design and experimental characterisation of a stable human heparanase variant.
Rsc Chem Biol, 3, 2022
|
|
7S4B
 
 | |
7S3S
 
 | |
7S3K
 
 | |
7T2V
 
 | |
7T2T
 
 | |
7T2U
 
 | |
7THM
 
 | | SARS-CoV-2 nsp12/7/8 complex with a native N-terminus nsp9 | | Descriptor: | MANGANESE (II) ION, Non-structural protein 7, Non-structural protein 8, ... | | Authors: | Osinski, A, Tagliabracci, V.S, Chen, Z, Li, Y. | | Deposit date: | 2022-01-11 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | The mechanism of RNA capping by SARS-CoV-2.
Nature, 609, 2022
|
|
7U0N
 
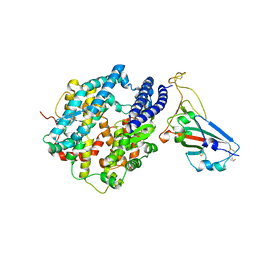 | | Crystal structure of chimeric omicron RBD (strain BA.1) complexed with human ACE2 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Geng, Q, Shi, K, Ye, G, Zhang, W, Aihara, H, Li, F. | | Deposit date: | 2022-02-18 | | Release date: | 2022-03-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structural Basis for Human Receptor Recognition by SARS-CoV-2 Omicron Variant BA.1.
J.Virol., 96, 2022
|
|
7KFI
 
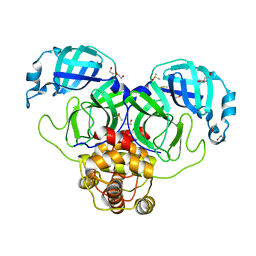 | | SARS-CoV-2 Main protease immature form - apo structure | | Descriptor: | 3C-like proteinase, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE | | Authors: | Noske, G.D, Nakamura, A.M, Gawriljuk, V.O, Lima, G.M.A, Zeri, A.C.M, Nascimento, A.F.Z, Oliva, G, Godoy, A.S. | | Deposit date: | 2020-10-14 | | Release date: | 2020-10-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A Crystallographic Snapshot of SARS-CoV-2 Main Protease Maturation Process.
J.Mol.Biol., 433, 2021
|
|
7L1F
 
 | | SARS-CoV-2 RdRp in complex with 4 Remdesivir monophosphate | | Descriptor: | Non-structural protein 7, Non-structural protein 8, RNA (5'-R(P*AP*UP*UP*UP*UP*AP*AP*UP*AP*GP*CP*UP*UP*CP*UP*UP*AP*G)-3'), ... | | Authors: | Bravo, J.P.K, Taylor, D.W. | | Deposit date: | 2020-12-14 | | Release date: | 2021-02-10 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.89 Å) | | Cite: | Remdesivir is a delayed translocation inhibitor of SARS-CoV-2 replication.
Mol.Cell, 81, 2021
|
|
7LDX
 
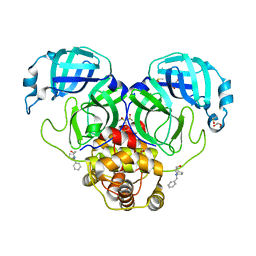 | | SARS-CoV-2 Main protease immature form - F2X Entry Library E06 fragment | | Descriptor: | (3-endo)-8-benzyl-8-azabicyclo[3.2.1]octan-3-ol, 3C-like proteinase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Noske, G.D, Nakamura, A.M, Gawriljuk, V.O, Lima, G.M.A, Zeri, A.C.M, Nascimento, A.F.Z, Oliva, G, Godoy, A.S. | | Deposit date: | 2021-01-14 | | Release date: | 2021-01-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | A Crystallographic Snapshot of SARS-CoV-2 Main Protease Maturation Process.
J.Mol.Biol., 433, 2021
|
|
7LFP
 
 | | SARS-CoV-2 Main protease immature form - F2X Entry Library G05 fragment | | Descriptor: | 3C-like proteinase, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Noske, G.D, Nakamura, A.M, Gawriljuk, V.O, Lima, G.M.A, Zeri, A.C.M, Nascimento, A.F.Z, Oliva, G, Godoy, A.S. | | Deposit date: | 2021-01-18 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Crystallographic Snapshot of SARS-CoV-2 Main Protease Maturation Process.
J.Mol.Biol., 433, 2021
|
|
7LFE
 
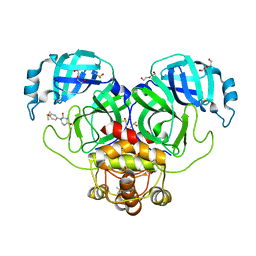 | | SARS-CoV-2 Main protease immature form - F2X Entry Library E03 fragment | | Descriptor: | (2R,4R)-1-phenylhexahydropyrimidine-2,4-diol, 3C-like proteinase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Noske, G.D, Nakamura, A.M, Gawriljuk, V.O, Lima, G.M.A, Zeri, A.C.M, Nascimento, A.F.Z, Oliva, G, Godoy, A.S. | | Deposit date: | 2021-01-16 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | A Crystallographic Snapshot of SARS-CoV-2 Main Protease Maturation Process.
J.Mol.Biol., 433, 2021
|
|
7JWB
 
 | |
7LUX
 
 | | AALALL segment from the Nucleoprotein of SARS-CoV-2, residues 217-222, crystal form 2 | | Descriptor: | Nucleoprotein AALALL, TETRAETHYLENE GLYCOL | | Authors: | Lu, J, Zee, C.-T, Sawaya, M.R, Rodriguez, J.A, Eisenberg, D.S. | | Deposit date: | 2021-02-23 | | Release date: | 2021-03-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.303 Å) | | Cite: | Inhibition of amyloid formation of the Nucleoprotein of SARS-CoV-2.
Biorxiv, 2021
|
|
7LUZ
 
 | |
7LV2
 
 | |
7LTU
 
 | | AALALL SEGMENT FROM THE NUCLEOPROTEIN OF SARS-COV-2, RESIDUES 217-222, CRYSTAL FORM 1 | | Descriptor: | AALALL SEGMENT FROM THE NUCLEOPROTEIN OF SARS-COV-2,RESIDUES 217-222, trifluoroacetic acid | | Authors: | Zee, C.-T, Sawaya, M.R, Rodriguez, J.A, Eisenberg, D.S. | | Deposit date: | 2021-02-20 | | Release date: | 2021-03-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.122 Å) | | Cite: | Inhibition of amyloid formation of the Nucleoprotein of SARS-CoV-2.
Biorxiv, 2021
|
|
7LJR
 
 | | SARS-CoV-2 Spike Protein Trimer bound to DH1043 fab | | Descriptor: | Fab DH1043 heavy chain, Fab DH1043 light chain, Spike glycoprotein | | Authors: | Gobeil, S, Acharya, P. | | Deposit date: | 2021-01-30 | | Release date: | 2021-03-31 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | The functions of SARS-CoV-2 neutralizing and infection-enhancing antibodies in vitro and in mice and nonhuman primates.
Biorxiv, 2021
|
|
7LX5
 
 | |
7CHF
 
 | |
7CHH
 
 | | Cryo-EM structure of the SARS-CoV-2 S-6P in complex with BD-368-2 Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BD-368-2 Fab heavy chain, ... | | Authors: | Xiao, J, Zhu, Q, Wang, G. | | Deposit date: | 2020-07-05 | | Release date: | 2020-09-16 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Structurally Resolved SARS-CoV-2 Antibody Shows High Efficacy in Severely Infected Hamsters and Provides a Potent Cocktail Pairing Strategy.
Cell, 183, 2020
|
|
