5DNY
 
 | | Structure of the ATPrS-Mre11/Rad50-DNA complex | | Descriptor: | DNA (27-MER), DNA double-strand break repair Rad50 ATPase,DNA double-strand break repair Rad50 ATPase, DNA double-strand break repair protein Mre11, ... | | Authors: | Liu, Y. | | Deposit date: | 2015-09-10 | | Release date: | 2016-02-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | ATP-dependent DNA binding, unwinding, and resection by the Mre11/Rad50 complex.
Embo J., 35, 2016
|
|
6WE1
 
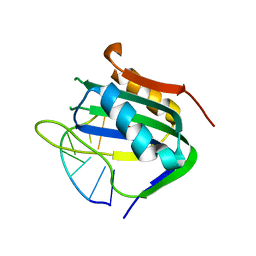 | | Wheat dwarf virus Rep domain complexed with a single-stranded DNA 8-mer comprising the cleavage site | | Descriptor: | DNA (5'-D(*AP*AP*TP*AP*TP*TP*AP*C)-3'), MANGANESE (II) ION, Replication-associated protein | | Authors: | Tompkins, K, Litzau, L.A, Pornschloegl, L, Nelson, A.T, Evans III, R.L, Gordon, W.R. | | Deposit date: | 2020-04-01 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.612 Å) | | Cite: | Molecular underpinnings of ssDNA specificity by Rep HUH-endonucleases and implications for HUH-tag multiplexing and engineering.
Nucleic Acids Res., 49, 2021
|
|
2XZF
 
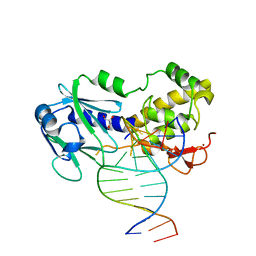 | | CRYSTAL STRUCTURE OF A COMPLEX BETWEEN THE WILD-TYPE LACTOCOCCUS LACTIS FPG (MUTM) AND AN OXIDIZED PYRIMIDINE CONTAINING DNA AT 293K | | Descriptor: | 5'-D(*CP*TP*CP*TP*TP*TP*VETP*TP*TP*TP*CP*TP*CP*GP)-3', 5'-D(*GP*AP*GP*AP*AP*AP*CP*AP*AP*AP*GP*AP*GP*CP)-3', FORMAMIDOPYRIMIDINE-DNA GLYCOSYLASE, ... | | Authors: | LeBihan, Y.V, Izquierdo, M.A, Coste, F, Culard, F, Gehrke, T.H, Essalhi, K, Aller, P, Carrel, T, Castaing, B. | | Deposit date: | 2010-11-25 | | Release date: | 2011-10-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | 5-Hydroxy-5-Methylhydantoin DNA Lesion, a Molecular Trap for DNA Glycosylases
Nucleic Acids Res., 39, 2011
|
|
4KB1
 
 | |
3MGH
 
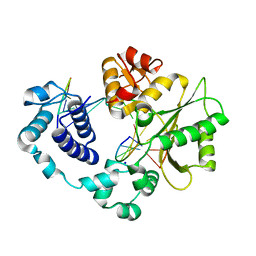 | | Binary complex of a DNA polymerase lambda loop mutant | | Descriptor: | DNA (5'-D(*CP*AP*GP*TP*AP*C)-3'), DNA (5'-D(*CP*GP*GP*CP*AP*GP*TP*AP*CP*TP*G)-3'), DNA (5'-D(P*GP*CP*CP*G)-3'), ... | | Authors: | Garcia-Diaz, M, Bebenek, K, Zhou, R.Z, Povirk, L.F, Kunkel, T. | | Deposit date: | 2010-04-06 | | Release date: | 2010-05-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Loop 1 modulates the fidelity of DNA polymerase lambda
Nucleic Acids Res., 38, 2010
|
|
6WE0
 
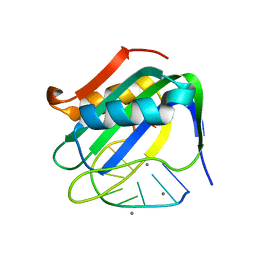 | | Wheat dwarf virus Rep domain complexed with a single-stranded DNA 10-mer comprising the cleavage site | | Descriptor: | DNA (5'-D(*TP*AP*AP*TP*AP*TP*TP*AP*CP*C)-3'), MANGANESE (II) ION, Replication-associated protein | | Authors: | Tompkins, K, Litzau, L.A, Shi, K, Nelson, A, Evans III, R.L, Gordon, W.R. | | Deposit date: | 2020-04-01 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular underpinnings of ssDNA specificity by Rep HUH-endonucleases and implications for HUH-tag multiplexing and engineering.
Nucleic Acids Res., 49, 2021
|
|
3VK8
 
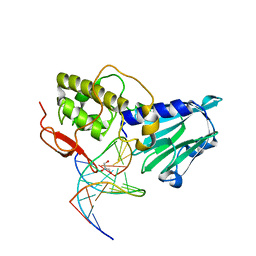 | | Crystal structure of DNA-glycosylase bound to DNA containing Thymine glycol | | Descriptor: | DNA (5'-D(*CP*GP*TP*CP*CP*AP*(CTG)P*GP*TP*CP*TP*AP*C)-3'), DNA (5'-D(*GP*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*G)-3'), GLYCEROL, ... | | Authors: | Imamura, K, Averill, A, Wallace, S.S, Doublie, S. | | Deposit date: | 2011-11-10 | | Release date: | 2011-12-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural characterization of viral ortholog of human DNA glycosylase NEIL1 bound to thymine glycol or 5-hydroxyuracil-containing DNA
J.Biol.Chem., 287, 2012
|
|
1OKB
 
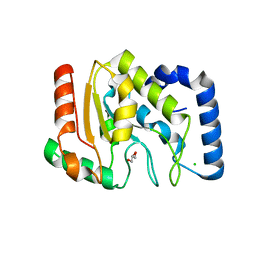 | | crystal structure of Uracil-DNA glycosylase from Atlantic cod (Gadus morhua) | | Descriptor: | CHLORIDE ION, GLYCEROL, URACIL-DNA GLYCOSYLASE | | Authors: | Leiros, I, Moe, E, Lanes, O, Smalas, A.O, Willassen, N.P. | | Deposit date: | 2003-07-21 | | Release date: | 2004-04-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Crystal Structure of Uracil-DNA Glycosylase from Atlantic Cod (Gadus Morhua) Reveals Cold-Adaptation Features
Acta Crystallogr.,Sect.D, 59, 2003
|
|
2J8X
 
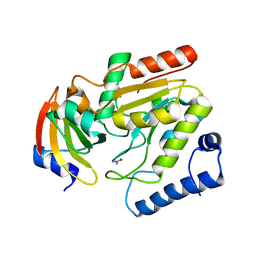 | | Epstein-Barr virus uracil-DNA glycosylase in complex with Ugi from PBS-2 | | Descriptor: | URACIL-DNA GLYCOSYLASE, URACIL-DNA GLYCOSYLASE INHIBITOR, UREA | | Authors: | Geoui, T, Buisson, M, Tarbouriech, N, Burmeister, W.P. | | Deposit date: | 2006-10-31 | | Release date: | 2006-12-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | New Insights on the Role of the Gamma-Herpesvirus Uracil-DNA Glycosylase Leucine Loop Revealed by the Structure of the Epstein-Barr Virus Enzyme in Complex with an Inhibitor Protein.
J.Mol.Biol., 366, 2007
|
|
3KOV
 
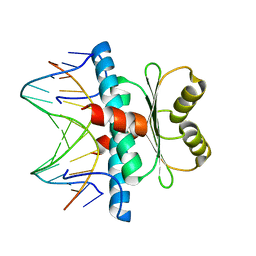 | | Structure of MEF2A bound to DNA reveals a completely folded MADS-box/MEF2 domain that recognizes DNA and recruits transcription co-factors | | Descriptor: | DNA (5'-D(*AP*AP*CP*TP*AP*TP*TP*TP*AP*TP*AP*AP*G)-3'), DNA (5'-D(*TP*CP*TP*TP*AP*TP*AP*AP*AP*TP*AP*GP*T)-3'), Myocyte-specific enhancer factor 2A | | Authors: | Wu, Y, Dey, R, Han, A, Jayathilaka, N, Philips, M, Ye, J, Chen, L. | | Deposit date: | 2009-11-14 | | Release date: | 2010-02-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of the MADS-box/MEF2 Domain of MEF2A Bound to DNA and Its Implication for Myocardin Recruitment.
J.Mol.Biol., 397, 2010
|
|
2LKX
 
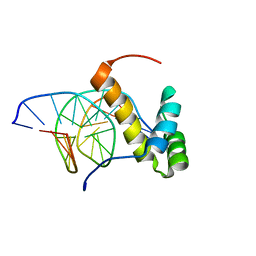 | | NMR structure of the homeodomain of Pitx2 in complex with a TAATCC DNA binding site | | Descriptor: | DNA (5'-D(*CP*GP*GP*GP*GP*AP*TP*TP*AP*GP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*CP*TP*AP*AP*TP*CP*CP*CP*CP*G)-3'), Pituitary homeobox 3 | | Authors: | Baird-Titus, J.M, Doerdelmann, T, Chaney, B.A, Clark-Baldwin, K, Dave, V, Ma, J. | | Deposit date: | 2011-10-21 | | Release date: | 2012-05-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the K50 class homeodomain PITX2 bound to DNA and implications for mutations that cause Rieger syndrome
Biochemistry, 44, 2005
|
|
8K3D
 
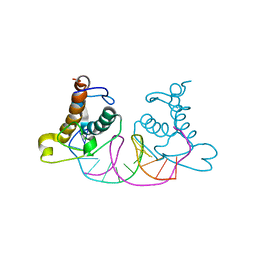 | | Crystal structure of NRF1 DBD bound to DNA | | Descriptor: | DNA (5'-D(*GP*GP*TP*GP*CP*GP*CP*AP*TP*GP*CP*GP*CP*AP*CP*C)-3'), Nuclear respiratory factor 1 | | Authors: | Li, W.F, Liu, K, Min, J.R. | | Deposit date: | 2023-07-15 | | Release date: | 2023-12-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular mechanism of specific DNA sequence recognition by NRF1.
Nucleic Acids Res., 52, 2024
|
|
8K4L
 
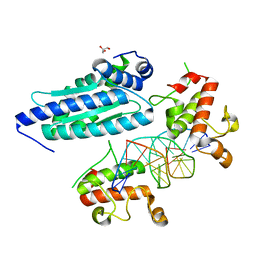 | |
6FB8
 
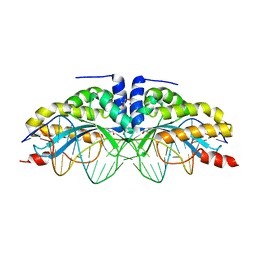 | |
6FB7
 
 | |
6CG8
 
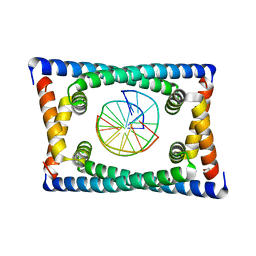 | | Structure of C. crescentus GapR-DNA | | Descriptor: | DNA (5'-D(*TP*TP*AP*AP*AP*AP*TP*TP*AP*AP*A)-3'), DNA (5'-D(*TP*TP*TP*AP*AP*TP*TP*TP*TP*AP*A)-3'), UPF0335 protein B7Z12_12435 | | Authors: | Schumacher, M.A. | | Deposit date: | 2018-02-19 | | Release date: | 2018-09-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.299 Å) | | Cite: | A Bacterial Chromosome Structuring Protein Binds Overtwisted DNA to Stimulate Type II Topoisomerases and Enable DNA Replication.
Cell, 175, 2018
|
|
8EWV
 
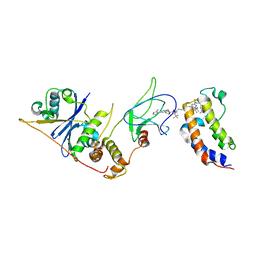 | | DNA-encoded library (DEL)-enabled discovery of proximity inducing small molecules | | Descriptor: | Bromodomain-containing protein 4, Elongin-B, Elongin-C, ... | | Authors: | Schreiber, S.L, Shu, W, Ma, X, Michaud, G, Bonazzi, S, Berst, F. | | Deposit date: | 2022-10-24 | | Release date: | 2023-10-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | DNA-encoded library-enabled discovery of proximity-inducing small molecules.
Nat.Chem.Biol., 20, 2024
|
|
161D
 
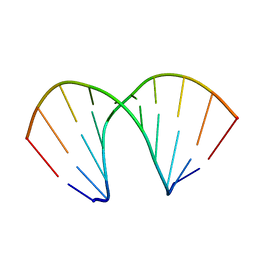 | |
1RFI
 
 | | Crystal structure of human Tyrosyl-DNA Phosphodiesterase complexed with vanadate, pentapeptide KLNYK, and tetranucleotide AGTC | | Descriptor: | 5'-D(*AP*GP*TP*C)-3', SPERMINE, Topoisomerase I-Derived Peptide, ... | | Authors: | Davies, D.R, Interthal, H, Champoux, J.J, Hol, W.G. | | Deposit date: | 2003-11-10 | | Release date: | 2004-03-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Explorations of peptide and oligonucleotide binding sites of tyrosyl-DNA phosphodiesterase using vanadate complexes.
J.Med.Chem., 47, 2004
|
|
8SVA
 
 | | Structure of the Rhodococcus sp. USK13 DarR-20 bp DNA complex | | Descriptor: | DNA (5'-D(*TP*AP*GP*AP*TP*AP*CP*TP*CP*CP*GP*GP*AP*GP*TP*AP*TP*CP*TP*A)-3'), PHOSPHATE ION, TetR/AcrR family transcriptional regulator | | Authors: | Schumacher, M.A. | | Deposit date: | 2023-05-15 | | Release date: | 2023-11-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Structures of the DarR transcription regulator reveal unique modes of second messenger and DNA binding.
Nat Commun, 14, 2023
|
|
1MU9
 
 | | Crystal Structure of a Human Tyrosyl-DNA Phosphodiesterase (Tdp1)-Vanadate Complex | | Descriptor: | GLYCEROL, Tyrosyl-DNA Phosphodiesterase, VANADATE ION | | Authors: | Davies, D.R, Interthal, H, Champoux, J.J, Hol, W.G.J. | | Deposit date: | 2002-09-23 | | Release date: | 2003-01-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Insights Into Substrate Binding and Catalytic Mechanism of Human Tyrosyl-DNA Phosphodiesterase (Tdp1) from Vanadate- and Tungstate-Inhibited Structures
J.Mol.Biol., 324, 2002
|
|
1GTC
 
 | | HUMAN IMMUNODEFICIENCY VIRUS-1 OKAZAKI FRAGMENT, DNA-RNA CHIMERA, NMR, 11 STRUCTURES | | Descriptor: | DNA (5'-D(*GP*CP*AP*GP*TP*GP*GP*C)-3'), DNA/RNA (5'-R(*GP*CP*CP*A)-D(P*CP*TP*GP*C)-3') | | Authors: | Fedoroff, O.Y, Salazar, M, Reid, B.R. | | Deposit date: | 1996-06-13 | | Release date: | 1996-12-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural variation among retroviral primer-DNA junctions: solution structure of the HIV-1 (-)-strand Okazaki fragment r(gcca)d(CTGC).d(GCAGTGGC).
Biochemistry, 35, 1996
|
|
4G4Q
 
 | | MutM containing F114A mutation bound to undamaged DNA | | Descriptor: | DNA (5'-D(*AP*GP*GP*TP*AP*GP*AP*CP*TP*CP*GP*GP*AP*CP*GP*C)-3'), DNA (5'-D(*TP*GP*CP*GP*TP*CP*CP*GP*AP*GP*(TX2)P*CP*TP*AP*CP*C)-3'), Formamidopyrimidine-DNA glycosylase, ... | | Authors: | Sung, R.J, Zhang, M, Qi, Y, Verdine, G.L. | | Deposit date: | 2012-07-16 | | Release date: | 2013-02-20 | | Last modified: | 2013-04-24 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural and Biochemical Analysis of DNA Helix Invasion by the Bacterial 8-Oxoguanine DNA Glycosylase MutM.
J.Biol.Chem., 288, 2013
|
|
1RFF
 
 | | Crystal structure of human Tyrosyl-DNA Phosphodiesterase complexed with vanadate, octapeptide KLNYYDPR, and tetranucleotide AGTT. | | Descriptor: | 5'-D(*AP*GP*TP*T)-3', SPERMINE, Topoisomerase I-Derived Peptide, ... | | Authors: | Davies, D.R, Interthal, H, Champoux, J.J, Hol, W.G. | | Deposit date: | 2003-11-08 | | Release date: | 2004-03-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Explorations of peptide and oligonucleotide binding sites of tyrosyl-DNA phosphodiesterase using vanadate complexes.
J.Med.Chem., 47, 2004
|
|
1RG2
 
 | | Crystal structure of human Tyrosyl-DNA Phosphodiesterase complexed with vanadate, octopamine, and tetranucleotide AGTA | | Descriptor: | 4-(2R-AMINO-1-HYDROXYETHYL)PHENOL, 4-(2S-AMINO-1-HYDROXYETHYL)PHENOL, 5'-D(*AP*GP*TP*A)-3', ... | | Authors: | Davies, D.R, Interthal, H, Champoux, J.J, Hol, W.G. | | Deposit date: | 2003-11-11 | | Release date: | 2004-03-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Explorations of peptide and oligonucleotide binding sites of tyrosyl-DNA phosphodiesterase using vanadate complexes.
J.Med.Chem., 47, 2004
|
|
