8CVM
 
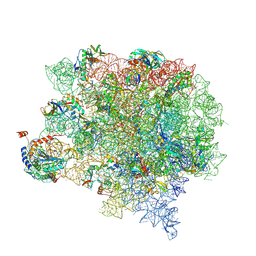 | | Cutibacterium acnes 50S ribosomal subunit with P-site tRNA and Sarecycline bound in the local refined map | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Lomakin, I.B, Devarkar, S.C, Bunick, C.G. | | Deposit date: | 2022-05-18 | | Release date: | 2023-03-15 | | Last modified: | 2023-04-19 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Sarecycline inhibits protein translation in Cutibacterium acnes 70S ribosome using a two-site mechanism.
Nucleic Acids Res., 51, 2023
|
|
7VCE
 
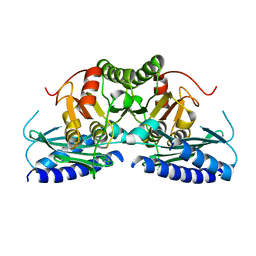 | | Structural studies of human inositol monophosphatase-1 inhibition by ebselen | | Descriptor: | Inositol monophosphatase 1 | | Authors: | Abuhammad, A, Laurieri, N, Rice, A, Lowe, E.D, McDonough, M.A, Singh, N, Churchill, G.C. | | Deposit date: | 2021-09-02 | | Release date: | 2022-09-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and biochemical analysis of human inositol monophosphatase-1 inhibition by ebselen.
J.Biomol.Struct.Dyn., 2023
|
|
8CXR
 
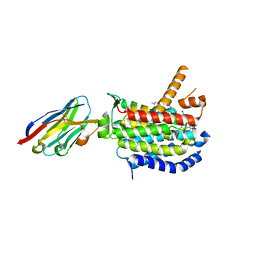 | | Crystal structure of MraY bound to a sphaerimicin analogue | | Descriptor: | (1S,4R,5S,6R,7S,9S,10S,11S,13S,14R)-9-[(2S,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxyoxolan-2-yl]-14-(hexadecanoyloxy)-5,6,13-trihydroxy-8,16-dioxa-2,11-diazatricyclo[9.3.1.1~4,7~]hexadecane-10-carboxylic acid, MraYAA nanobody, Phospho-N-acetylmuramoyl-pentapeptide-transferase | | Authors: | Mashalidis, E.H, Lee, S.Y. | | Deposit date: | 2022-05-22 | | Release date: | 2023-03-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | Synthesis of macrocyclic nucleoside antibacterials and their interactions with MraY.
Nat Commun, 13, 2022
|
|
6LLZ
 
 | |
8D7R
 
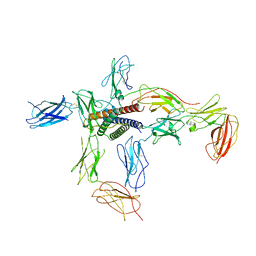 | |
8D74
 
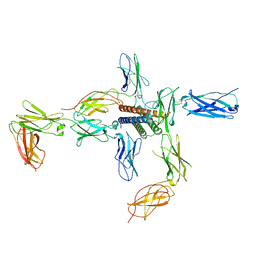 | |
8DFM
 
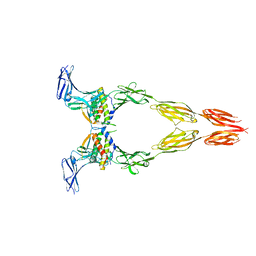 | | Ectodomain of full-length wild-type KIT-SCF dimers | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Isoform 2 of Mast/stem cell growth factor receptor Kit, ... | | Authors: | Krimmer, S.G, Bertoletti, N, Mi, W, Schlessinger, J. | | Deposit date: | 2022-06-22 | | Release date: | 2023-03-29 | | Last modified: | 2023-04-05 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Cryo-EM analyses of KIT and oncogenic mutants reveal structural oncogenic plasticity and a target for therapeutic intervention.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8D6A
 
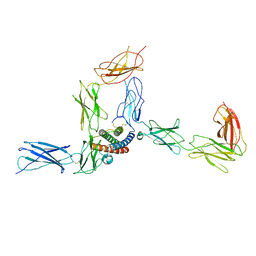 | |
7VOZ
 
 | |
6M17
 
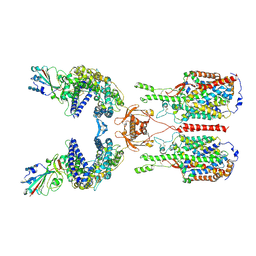 | | The 2019-nCoV RBD/ACE2-B0AT1 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yan, R.H, Zhang, Y.Y, Li, Y.N, Xia, L, Guo, Y.Y, Zhou, Q. | | Deposit date: | 2020-02-24 | | Release date: | 2020-03-11 | | Last modified: | 2021-03-10 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2.
Science, 367, 2020
|
|
7VIB
 
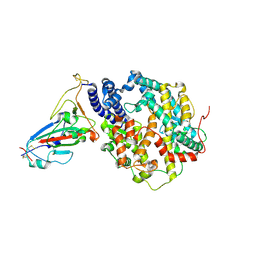 | | Crystal structure of human ACE2 and GX/P2V RBD | | Descriptor: | Angiotensin-converting enzyme 2, Spike glycoprotein, ZINC ION | | Authors: | Guo, Y, Cao, W, Jia, N, Wang, W, Yuan, S, Wang, Y. | | Deposit date: | 2021-09-26 | | Release date: | 2022-10-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of human ACE2 and GX/P2V RBD
To Be Published
|
|
8D85
 
 | |
7VAA
 
 | |
8D82
 
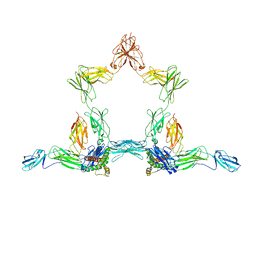 | |
6M49
 
 | | cryo-EM structure of Scap/Insig complex in the present of 25-hydroxyl cholesterol. | | Descriptor: | 25-HYDROXYCHOLESTEROL, Insulin-induced gene 2 protein, Sterol regulatory element-binding protein cleavage-activating protein,Sterol regulatory element-binding protein cleavage-activating protein | | Authors: | Yan, R, Cao, P, Song, W, Qian, H, Du, X, Coates, H.W, Zhao, X, Li, Y, Gao, S, Gong, X, Liu, X, Sui, J, Lei, J, Yang, H, Brown, A.J, Zhou, Q, Yan, C, Yan, N. | | Deposit date: | 2020-03-06 | | Release date: | 2021-01-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | A structure of human Scap bound to Insig-2 suggests how their interaction is regulated by sterols.
Science, 371, 2021
|
|
7VA8
 
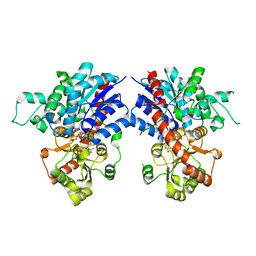 | | Crystal structure of MiCGT | | Descriptor: | UDP-glycosyltransferase 13, URIDINE-5'-DIPHOSPHATE-GLUCOSE | | Authors: | Zhong, L, Zhang, Z.M. | | Deposit date: | 2021-08-27 | | Release date: | 2022-07-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.85003233 Å) | | Cite: | Directed Evolution of a Plant Glycosyltransferase for Chemo- and Regioselective Glycosylation of Pharmaceutically Significant Flavonoids
Acs Catalysis, 11, 2021
|
|
8DFP
 
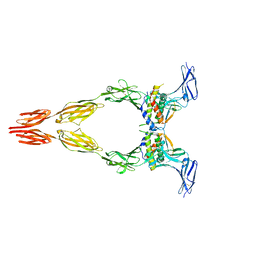 | | Ectodomain of full-length KIT(DupA502,Y503)-SCF dimers | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Isoform 2 of Mast/stem cell growth factor receptor Kit, ... | | Authors: | Bertoletti, N, Krimmer, S.G, Mi, W, Schlessinger, J. | | Deposit date: | 2022-06-22 | | Release date: | 2023-03-29 | | Last modified: | 2023-04-05 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Cryo-EM analyses of KIT and oncogenic mutants reveal structural oncogenic plasticity and a target for therapeutic intervention.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8DHA
 
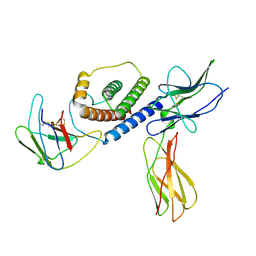 | |
8DH8
 
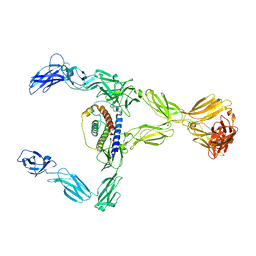 | |
8DH9
 
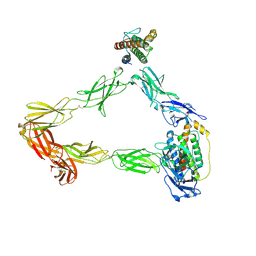 | |
6M67
 
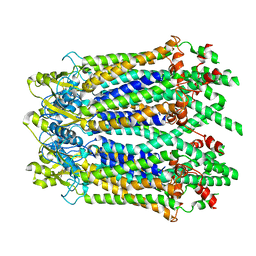 | | The Cryo-EM Structure of Human Pannexin 1 with D376E/D379E Mutation | | Descriptor: | Pannexin-1 | | Authors: | Jin, Q, Bo, Z, Xiang, Z, Xiaokang, Z, Ye, S. | | Deposit date: | 2020-03-13 | | Release date: | 2020-04-15 | | Last modified: | 2020-05-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structures of human pannexin 1 channel.
Cell Res., 30, 2020
|
|
8EUC
 
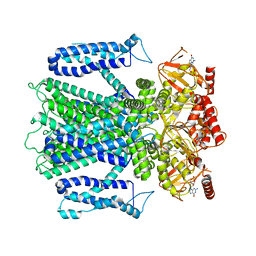 | | Cryo-EM structure of cGMP bound human CNGA3/CNGB3 channel in GDN, transition state 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CYCLIC GUANOSINE MONOPHOSPHATE, Cyclic nucleotide-gated cation channel alpha-3, ... | | Authors: | Hu, Z, Zheng, X, Yang, J. | | Deposit date: | 2022-10-18 | | Release date: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (3.61 Å) | | Cite: | Conformational trajectory of allosteric gating of the human cone photoreceptor cyclic nucleotide-gated channel.
Nat Commun, 14, 2023
|
|
8EVA
 
 | |
8EVB
 
 | |
8EV9
 
 | |
