2XB9
 
 | | Structure of Helicobacter pylori type II dehydroquinase in complex with inhibitor compound (2R)-2-(4-methoxybenzyl)-3-dehydroquinic acid | | Descriptor: | (1R,2R,4S,5R)-1,4,5-TRIHYDROXY-2-(4-METHOXYBENZYL)-3-OXOCYCLOHEXANECARBOXYLIC ACID, 3-DEHYDROQUINATE DEHYDRATASE, CITRIC ACID | | Authors: | Otero, J.M, Tizon, L, Llamas-Saiz, A.L, Fox, G.C, Gonzalez-Bello, C, van Raaij, M.J. | | Deposit date: | 2010-04-08 | | Release date: | 2010-09-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Understanding the Key Factors that Control the Inhibition of Type II Dehydroquinase by (2R)-2- Benzyl-3-Dehydroquinic Acids.
Chemmedchem, 5, 2010
|
|
2Y5Z
 
 | | Mixed-function P450 MycG in complex with mycinamicin III in C2221 space group | | Descriptor: | BENZAMIDINE, GLYCEROL, MYCINAMICIN III, ... | | Authors: | Li, S, Kells, P.M, Sherman, D.H, Podust, L.M. | | Deposit date: | 2011-01-19 | | Release date: | 2012-02-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Substrate Recognition by the Multifunctional Cytochrome P450 Mycg in Mycinamicin Hydroxylation and Epoxidation Reactions.
J.Biol.Chem., 287, 2012
|
|
2XMS
 
 | | Crystal structure of human NDRG2 protein provides insight into its role as a tumor suppressor | | Descriptor: | CHLORIDE ION, IMIDAZOLE, PROTEIN NDRG2 | | Authors: | Hwang, J, Kim, Y, Lee, H, Kim, M.H. | | Deposit date: | 2010-07-29 | | Release date: | 2011-01-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structure of Human Ndrg2 Protein Provides Insight Into its Role as a Tumor Suppressor.
J.Biol.Chem., 286, 2011
|
|
1I5J
 
 | |
2XO5
 
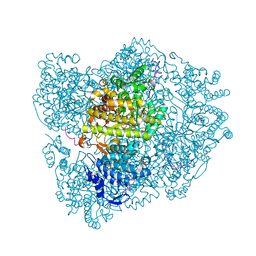 | | RIBONUCLEOTIDE REDUCTASE Y731NH2Y MODIFIED R1 SUBUNIT OF E. COLI | | Descriptor: | RIBONUCLEOSIDE-DIPHOSPHATE REDUCTASE 1 SUBUNIT ALPHA, RIBONUCLEOSIDE-DIPHOSPHATE REDUCTASE 1 SUBUNIT BETA | | Authors: | Minnihan, E.C, Seyedsayamdost, M.R, Uhlin, U, Stubbe, J. | | Deposit date: | 2010-08-09 | | Release date: | 2010-08-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Kinetics of Radical Intermediate Formation and Deoxynucleotide Production in 3-Aminotyrosine- Substituted Escherichia Coli Ribonucleotide Reductases.
J.Am.Chem.Soc., 133, 2011
|
|
4F1K
 
 | | Crystal structure of the MG2+ free VWA domain of plasmodium falciparum trap protein | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, SULFATE ION, ... | | Authors: | Pihlajamaa, T, Knuuti, J, Kajander, T, Sharma, A, Permi, P. | | Deposit date: | 2012-05-07 | | Release date: | 2013-01-30 | | Last modified: | 2013-05-22 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structure of Plasmodium falciparum TRAP (thrombospondin-related anonymous protein) A domain highlights distinct features in apicomplexan von Willebrand factor A homologues.
Biochem.J., 450, 2013
|
|
2XG8
 
 | |
2XMR
 
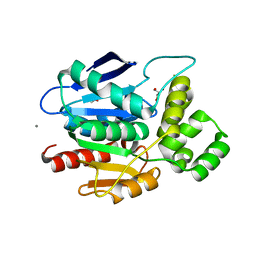 | | Crystal structure of human NDRG2 protein provides insight into its role as a tumor suppressor | | Descriptor: | ACETATE ION, CALCIUM ION, GLYCEROL, ... | | Authors: | Hwang, J, Kim, Y, Lee, H, Kim, M.H. | | Deposit date: | 2010-07-29 | | Release date: | 2011-01-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Human Ndrg2 Protein Provides Insight Into its Role as a Tumor Suppressor.
J.Biol.Chem., 286, 2011
|
|
1I6F
 
 | |
2XKP
 
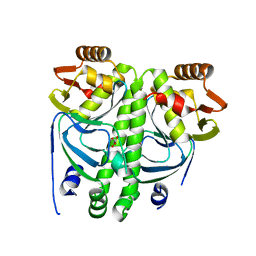 | |
1I7X
 
 | | BETA-CATENIN/E-CADHERIN COMPLEX | | Descriptor: | BETA-CATENIN, EPITHELIAL-CADHERIN | | Authors: | Huber, A.H, Weis, W.I. | | Deposit date: | 2001-03-10 | | Release date: | 2001-05-16 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structure of the beta-catenin/E-cadherin complex and the molecular basis of diverse ligand recognition by beta-catenin.
Cell(Cambridge,Mass.), 105, 2001
|
|
1IAH
 
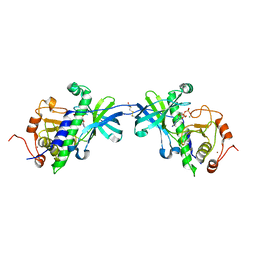 | | CRYSTAL STRUCTURE OF THE ATYPICAL PROTEIN KINASE DOMAIN OF A TRP CA-CHANNEL, CHAK (ADP-MG COMPLEX) | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Yamaguchi, H, Matsushita, M, Nairn, A.C, Kuriyan, J. | | Deposit date: | 2001-03-22 | | Release date: | 2001-06-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the atypical protein kinase domain of a TRP channel with phosphotransferase activity.
Mol.Cell, 7, 2001
|
|
1I8P
 
 | |
1IBR
 
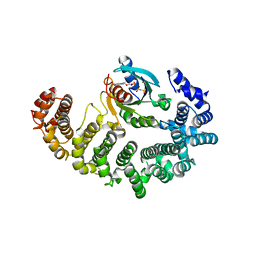 | | COMPLEX OF RAN WITH IMPORTIN BETA | | Descriptor: | GTP-binding nuclear protein RAN, Importin beta-1 subunit, MAGNESIUM ION, ... | | Authors: | Vetter, I.R, Arndt, A, Kutay, U, Goerlich, D, Wittinghofer, A. | | Deposit date: | 1999-05-14 | | Release date: | 1999-06-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural view of the Ran-Importin beta interaction at 2.3 A resolution
Cell(Cambridge,Mass.), 97, 1999
|
|
4XP4
 
 | | X-ray structure of Drosophila dopamine transporter in complex with cocaine | | Descriptor: | 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, ANTIBODY FRAGMENT HEAVY CHAIN-PROTEIN, 9D5-HEAVY CHAIN, ... | | Authors: | Aravind, P, Wang, K, Gouaux, E. | | Deposit date: | 2015-01-16 | | Release date: | 2015-05-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Neurotransmitter and psychostimulant recognition by the dopamine transporter.
Nature, 521, 2015
|
|
1I1R
 
 | | CRYSTAL STRUCTURE OF A CYTOKINE/RECEPTOR COMPLEX | | Descriptor: | INTERLEUKIN-6 RECEPTOR BETA CHAIN, VIRAL IL-6 | | Authors: | Chow, D, He, X, Snow, A.L, Rose-John, S, Garcia, K.C. | | Deposit date: | 2001-02-02 | | Release date: | 2001-03-28 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of an extracellular gp130 cytokine receptor signaling complex.
Science, 291, 2001
|
|
4FFO
 
 | | PylC in complex with phosphorylated D-ornithine | | Descriptor: | (2R)-2,5-diaminopentanoyl dihydrogen phosphate, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Quitterer, F, List, A, Beck, P, Bacher, A, Groll, M. | | Deposit date: | 2012-06-01 | | Release date: | 2012-09-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Biosynthesis of the 22nd genetically encoded amino acid pyrrolysine: structure and reaction mechanism of PylC at 1.5A resolution.
J.Mol.Biol., 424, 2012
|
|
1I52
 
 | | CRYSTAL STRUCTURE OF 4-DIPHOSPHOCYTIDYL-2-C-METHYLERYTHRITOL (CDP-ME) SYNTHASE (YGBP) INVOLVED IN MEVALONATE INDEPENDENT ISOPRENOID BIOSYNTHESIS | | Descriptor: | 4-DIPHOSPHOCYTIDYL-2-C-METHYLERYTHRITOL SYNTHASE, CALCIUM ION, CYTIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Richard, S.B, Bowman, M.E, Kwiatkowski, W, Kang, I, Chow, C, Lillo, M, Cane, D.E, Noel, J.P. | | Deposit date: | 2001-02-23 | | Release date: | 2001-07-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of 4-diphosphocytidyl-2-C- methylerythritol synthetase involved in mevalonate- independent isoprenoid biosynthesis.
Nat.Struct.Biol., 8, 2001
|
|
8F25
 
 | |
1I69
 
 | | CRYSTAL STRUCTURE OF THE REDUCED FORM OF OXYR | | Descriptor: | BENZOIC ACID, HYDROGEN PEROXIDE-INDUCIBLE GENES ACTIVATOR | | Authors: | Choi, H, Kim, S, Ryu, S. | | Deposit date: | 2001-03-02 | | Release date: | 2001-09-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of the redox switch in the OxyR transcription factor.
Cell(Cambridge,Mass.), 105, 2001
|
|
1I8G
 
 | | SOLUTION STRUCTURE OF PIN1 WW DOMAIN COMPLEXED WITH CDC25 PHOSPHOTHREONINE PEPTIDE | | Descriptor: | M-PHASE INDUCER PHOSPHATASE 3, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE NIMA-INTERACTING 1 | | Authors: | Wintjens, R, Wieruszeski, J.-M, Drobecq, H, Lippens, G, Landrieu, I. | | Deposit date: | 2001-03-14 | | Release date: | 2001-07-18 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | 1H NMR study on the binding of Pin1 Trp-Trp domain with phosphothreonine peptides.
J.Biol.Chem., 276, 2001
|
|
4FAP
 
 | |
4XPF
 
 | | X-ray structure of Drosophila dopamine transporter with subsiteB mutations (D121G/S426M) bound to RTI-55 | | Descriptor: | 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, ANTIBODY FRAGMENT HEAVY CHAIN-PROTEIN, 9D5-HEAVY CHAIN, ... | | Authors: | Penmatsa, A, Wang, K.H, Gouaux, E. | | Deposit date: | 2015-01-16 | | Release date: | 2015-05-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.273 Å) | | Cite: | Neurotransmitter and psychostimulant recognition by the dopamine transporter.
Nature, 521, 2015
|
|
2XO4
 
 | | RIBONUCLEOTIDE REDUCTASE Y730NH2Y MODIFIED R1 SUBUNIT OF E. COLI | | Descriptor: | RIBONUCLEOSIDE-DIPHOSPHATE REDUCTASE 1 SUBUNIT ALPHA, RIBONUCLEOSIDE-DIPHOSPHATE REDUCTASE 1 SUBUNIT BETA | | Authors: | Minnihan, E.C, Seyedsayamdost, M.R, Uhlin, U, Stubbe, J. | | Deposit date: | 2010-08-09 | | Release date: | 2010-08-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Kinetics of Radical Intermediate Formation and Deoxynucleotide Production in 3-Aminotyrosine- Substituted Escherichia Coli Ribonucleotide Reductases.
J.Am.Chem.Soc., 133, 2011
|
|
1I9B
 
 | | X-RAY STRUCTURE OF ACETYLCHOLINE BINDING PROTEIN (ACHBP) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETYLCHOLINE BINDING PROTEIN, CALCIUM ION | | Authors: | Brejc, K, van Dijk, W.J, Klaassen, R, Schuurmans, M, van der Oost, J, Smit, A.B, Sixma, T.K. | | Deposit date: | 2001-03-18 | | Release date: | 2001-05-16 | | Last modified: | 2018-03-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of an ACh-binding protein reveals the ligand-binding domain of nicotinic receptors.
Nature, 411, 2001
|
|
