4WWO
 
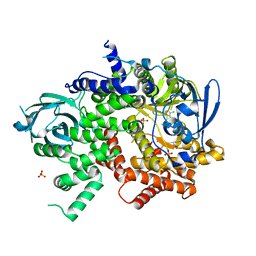 | | Crystal structure of human PI3K-gamma in complex with phenylquinoline inhibitor N-{(1S)-1-[8-chloro-2-(3-fluorophenyl)quinolin-3-yl]ethyl}-9H-purin-6-amine | | Descriptor: | N-{(1S)-1-[8-chloro-2-(3-fluorophenyl)quinolin-3-yl]ethyl}-9H-purin-6-amine, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION | | Authors: | Whittington, D.A, Tang, J, Yakowec, P. | | Deposit date: | 2014-11-11 | | Release date: | 2014-12-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery and in Vivo Evaluation of (S)-N-(1-(7-Fluoro-2-(pyridin-2-yl)quinolin-3-yl)ethyl)-9H-purin-6-amine (AMG319) and Related PI3K delta Inhibitors for Inflammation and Autoimmune Disease.
J.Med.Chem., 58, 2015
|
|
2AZ8
 
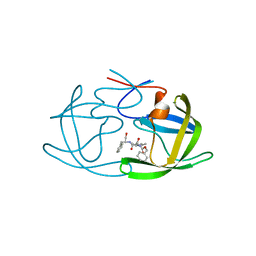 | | HIV-1 Protease NL4-3 in complex with inhibitor, TL-3 | | Descriptor: | PROTEASE RETROPEPSIN, benzyl [(1S,4S,7S,8R,9R,10S,13S,16S)-7,10-dibenzyl-8,9-dihydroxy-1,16-dimethyl-4,13-bis(1-methylethyl)-2,5,12,15,18-pentaoxo-20-phenyl-19-oxa-3,6,11,14,17-pentaazaicos-1-yl]carbamate | | Authors: | Heaslet, H, Kutilek, V, Morris, G.M, Lin, Y.-C, Elder, J.H, Torbett, B.E, Stout, C.D. | | Deposit date: | 2005-09-09 | | Release date: | 2006-02-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Insights into the Mechanisms of Drug Resistance in HIV-1 Protease NL4-3
J.Mol.Biol., 356, 2006
|
|
6DGY
 
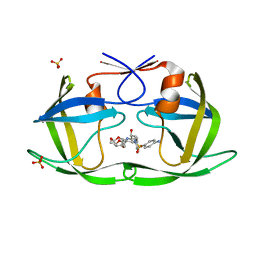 | | Crystal structure of HIV-1 Protease NL4-3 WT in complex with UMass1 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(1S,2R)-3-{[(4-aminophenyl)sulfonyl][(2S)-2-methylbutyl]amino}-1-benzyl-2-hydroxypropyl]carbamate, Protease, SULFATE ION | | Authors: | Lockbaum, G.J, Schiffer, C.A. | | Deposit date: | 2018-05-18 | | Release date: | 2018-12-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.954 Å) | | Cite: | Structural Adaptation of Darunavir Analogues against Primary Mutations in HIV-1 Protease.
ACS Infect Dis, 5, 2019
|
|
6DH7
 
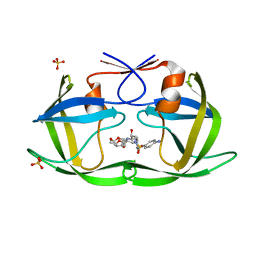 | | Crystal structure of HIV-1 Protease NL4-3 I50V Mutant in complex with UMass1 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(1S,2R)-3-{[(4-aminophenyl)sulfonyl][(2S)-2-methylbutyl]amino}-1-benzyl-2-hydroxypropyl]carbamate, Protease, SULFATE ION | | Authors: | Lockbaum, G.J, Schiffer, C.A. | | Deposit date: | 2018-05-18 | | Release date: | 2018-12-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Structural Adaptation of Darunavir Analogues against Primary Mutations in HIV-1 Protease.
ACS Infect Dis, 5, 2019
|
|
4YQU
 
 | | Glutathione S-transferase Omega 1 bound to covalent inhibitor C1-31 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Glutathione S-transferase omega-1, N-{5-(azepan-1-ylsulfonyl)-2-[(ethylsulfanyl)methoxy]phenyl}acetamide | | Authors: | Stuckey, J.A. | | Deposit date: | 2015-03-13 | | Release date: | 2016-10-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Mechanistic evaluation and transcriptional signature of a glutathione S-transferase omega 1 inhibitor.
Nat Commun, 7, 2016
|
|
6BI9
 
 | | Crystal Structure of Purine Nucleoside Phosphorylase Isoform 2 from Schistosoma mansoni in complex with 1,2,5-trimethyl-1H-pyrrole-3-carboxylic acid | | Descriptor: | 1,2,5-trimethyl-1H-pyrrole-3-carboxylic acid, DIMETHYL SULFOXIDE, Purine nucleoside phosphorylase | | Authors: | Faheem, M, Neto, J.B, Collins, P, Pearce, N.M, Valadares, N.F, Bird, L, Pereira, H.M, Delft, F.V, Barbosa, J.A.R.G. | | Deposit date: | 2017-11-01 | | Release date: | 2018-11-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Crystal Structure of Purine Nucleoside Phosphorylase Isoform 2 from Schistosoma mansoni in complex with 1,2,5-trimethyl-1H-pyrrole-3-carboxylic acid
To Be Published
|
|
6DH4
 
 | | Crystal structure of HIV-1 Protease NL4-3 V82I Mutant in complex with UMass1 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(1S,2R)-3-{[(4-aminophenyl)sulfonyl][(2S)-2-methylbutyl]amino}-1-benzyl-2-hydroxypropyl]carbamate, Protease, SULFATE ION | | Authors: | Lockbaum, G.J, Schiffer, C.A. | | Deposit date: | 2018-05-18 | | Release date: | 2018-12-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.943 Å) | | Cite: | Structural Adaptation of Darunavir Analogues against Primary Mutations in HIV-1 Protease.
ACS Infect Dis, 5, 2019
|
|
4ZA9
 
 | | Structure of A. niger fdc1 in complex with a phenylpyruvate derived adduct to the prenylated flavin cofactor | | Descriptor: | 1-deoxy-5-O-phosphono-1-[(1S)-3,3,4,5-tetramethyl-9,11-dioxo-1-(phenylacetyl)-2,3,8,9,10,11-hexahydro-1H,7H-quinolino[1 ,8-fg]pteridin-7-yl]-D-ribitol, MANGANESE (II) ION, POTASSIUM ION, ... | | Authors: | Payne, K.A.P, Leys, D. | | Deposit date: | 2015-04-13 | | Release date: | 2015-06-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.01 Å) | | Cite: | New cofactor supports alpha , beta-unsaturated acid decarboxylation via 1,3-dipolar cycloaddition.
Nature, 522, 2015
|
|
2AID
 
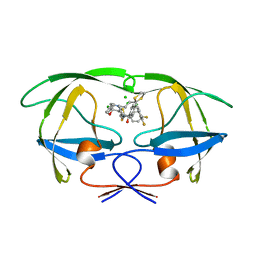 | | STRUCTURE OF A NON-PEPTIDE INHIBITOR COMPLEXED WITH HIV-1 PROTEASE: DEVELOPING A CYCLE OF STRUCTURE-BASED DRUG DESIGN | | Descriptor: | 4-(4-CHLORO-PHENYL)-1-{3-[2-(4-FLUORO-PHENYL)-[1,3]DITHIOLAN-2-YL]-PROPYL}-PIPERIDIN-4-OL, CHLORIDE ION, HUMAN IMMUNODEFICIENCY VIRUS PROTEASE | | Authors: | Rutenber, E.E, Fauman, E.B, Keenan, R.J, Stroud, R.M. | | Deposit date: | 1997-04-17 | | Release date: | 1997-10-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of a non-peptide inhibitor complexed with HIV-1 protease. Developing a cycle of structure-based drug design.
J.Biol.Chem., 268, 1993
|
|
5WLO
 
 | | a novel 13-ring macrocyclic HIV-1 protease inhibitors involving the P1'-P2' ligands | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl {(2S,3R)-3-hydroxy-4-[(7E)-13-methoxy-1,1-dioxo-1,4,5,6,9,11-hexahydro-10,1lambda~6~,2-benzoxathiazacyclotridecin-2(3H)-yl]-1-phenylbutan-2-yl}carbamate, ACETATE ION, CHLORIDE ION, ... | | Authors: | Wang, Y.-F, Agniswamy, J, Weber, I.T. | | Deposit date: | 2017-07-27 | | Release date: | 2017-10-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Design, synthesis, X-ray studies, and biological evaluation of novel macrocyclic HIV-1 protease inhibitors involving the P1'-P2' ligands.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
7EO4
 
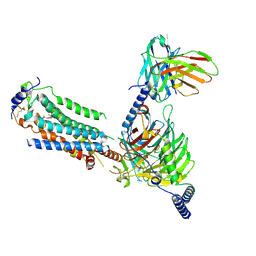 | | Cryo-EM of Sphingosine 1-phosphate receptor 1 / Gi complex bound to BAF312 | | Descriptor: | 1-[[4-[(~{E})-~{N}-[[4-cyclohexyl-3-(trifluoromethyl)phenyl]methoxy]-~{C}-methyl-carbonimidoyl]-2-ethyl-phenyl]methyl]azetidine-3-carboxylic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | He, Y, Xu, Z, Ikuta, T, Inoue, A. | | Deposit date: | 2021-04-21 | | Release date: | 2022-01-05 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Structural basis of sphingosine-1-phosphate receptor 1 activation and biased agonism.
Nat.Chem.Biol., 18, 2022
|
|
4YE3
 
 | | Crystal Structure of Multidrug Resistant HIV-1 Protease Clinical Isolate PR20 with Inhibitor GRL-4410A | | Descriptor: | (3R,3aS,4R,6aR)-4-methoxyhexahydrofuro[2,3-b]furan-3-yl [(2S,3R)-3-hydroxy-4-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}-1-phenylbutan-2-yl]carbamate, CHLORIDE ION, GLYCEROL, ... | | Authors: | Agniswamy, J, Weber, I.T. | | Deposit date: | 2015-02-23 | | Release date: | 2015-06-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Substituted Bis-THF Protease Inhibitors with Improved Potency against Highly Resistant Mature HIV-1 Protease PR20.
J.Med.Chem., 58, 2015
|
|
2YGQ
 
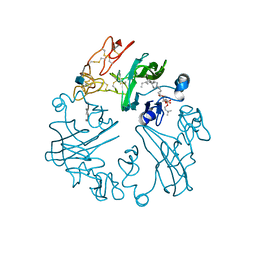 | | WIF domain-epidermal growth factor (EGF)-like domains 1-3 of human Wnt inhibitory factor 1 in complex with 1,2- dipalmitoylphosphatidylcholine | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSHOCHOLINE, 1,3,4,6-tetra-O-sulfo-beta-D-fructofuranose-(2-1)-2,3,4,6-tetra-O-sulfonato-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Malinauskas, T, Aricescu, A.R, Lu, W, Siebold, C, Jones, E.Y. | | Deposit date: | 2011-04-19 | | Release date: | 2011-07-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.951 Å) | | Cite: | Modular Mechanism of Wnt Signaling Inhibition by Wnt Inhibitory Factor 1
Nat.Struct.Mol.Biol., 18, 2011
|
|
6M8Z
 
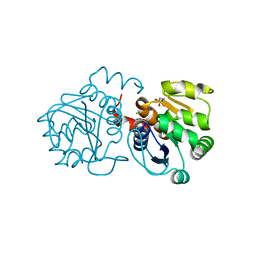 | | Crystal structure of human DJ-1 without a modification on Cys-106 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, Protein/nucleic acid deglycase DJ-1 | | Authors: | Shumilin, I.A, Shabalin, I.G, Shumilina, S.V, Werenskjold, C, Utepbergenov, D, Minor, W. | | Deposit date: | 2018-08-22 | | Release date: | 2018-09-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | A transient post-translational modification of active site cysteine alters binding properties of the parkinsonism protein DJ-1.
Biochem. Biophys. Res. Commun., 504, 2018
|
|
4YOA
 
 | | Crsystal structure HIV-1 Protease MDR769 L33F Complexed with darunavir | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, HIV-1 Protease | | Authors: | Kuiper, B.D, Keusch, B, Dewdney, T.G, Chordia, P, Brunzelle, J.S, Ross, K, Kovari, I.A, MacArthur, R, Salimnia, H, Kovari, L.C. | | Deposit date: | 2015-03-11 | | Release date: | 2015-07-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.697 Å) | | Cite: | The L33F darunavir resistance mutation acts as a molecular anchor reducing the flexibility of the HIV-1 protease 30s and 80s loops.
Biochem Biophys Rep, 2, 2015
|
|
6MHB
 
 | | Glutathione S-Transferase Omega 1 bound to covalent inhibitor 18 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Glutathione S-transferase omega-1, N-[4-(4-chlorophenyl)-1,3-thiazol-2-yl]propanamide | | Authors: | Petrunak, E.M, Stuckey, J.A. | | Deposit date: | 2018-09-17 | | Release date: | 2019-02-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure-Based Design of N-(5-Phenylthiazol-2-yl)acrylamides as Novel and Potent Glutathione S-Transferase Omega 1 Inhibitors.
J. Med. Chem., 62, 2019
|
|
4R5P
 
 | | Crystal structure of HIV-1 reverse transcriptase (RT) with DNA and a nucleoside triphosphate mimic alpha-carboxy nucleoside phosphonate inhibitor | | Descriptor: | 5'-D(*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*G)-3', 5'-D(*TP*GP*GP*AP*CP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*G)-3', HIV-1 reverse transcriptase, ... | | Authors: | Das, K, Martinez, S.E, Arnold, E. | | Deposit date: | 2014-08-21 | | Release date: | 2015-03-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.894 Å) | | Cite: | Alpha-carboxy nucleoside phosphonates as universal nucleoside triphosphate mimics.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
3HR9
 
 | | The truncated Fibrobacter succinogenes 1,3-1,4-beta-D-glucanase F40I mutant | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, Beta-glucanase, ... | | Authors: | Tsai, L.C, Huang, H.C, Hsiao, C.H. | | Deposit date: | 2009-06-09 | | Release date: | 2009-07-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The truncated Fibrobacter succinogenes 1,3-1,4-beta-D-glucanase mutant F40I
To be Published
|
|
4ZGG
 
 | |
4XOJ
 
 | | Structure of bovine trypsin in complex with analogues of sunflower inhibitor 1 (SFTI-1) | | Descriptor: | 1,2-ETHANEDIOL, AMMONIUM ION, CALCIUM ION, ... | | Authors: | Golik, P, Malicki, S, Grudnik, P, Karna, N, Debowski, D, Legowska, A, Wladyka, B, Gitlin, A, Brzozowski, K, Dubin, G, Rolka, K. | | Deposit date: | 2015-01-16 | | Release date: | 2015-08-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (0.91 Å) | | Cite: | Investigation of Serine-Proteinase-Catalyzed Peptide Splicing in Analogues of Sunflower Trypsin Inhibitor 1 (SFTI-1).
Chembiochem, 16, 2015
|
|
5UEM
 
 | | Crystal structure of 354NC37 Fab in complex with HIV-1 clade AE strain 93TH057 gp120 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 354NC37 Fab Heavy Chain, ... | | Authors: | Sievers, S.A, Gristick, H.B, Bjorkman, P.J. | | Deposit date: | 2017-01-02 | | Release date: | 2018-01-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Coexistence of potent HIV-1 broadly neutralizing antibodies and antibody-sensitive viruses in a viremic controller.
Sci Transl Med, 9, 2017
|
|
6DH3
 
 | | Crystal structure of HIV-1 Protease NL4-3 V82I Mutant in complex with darunavir | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, Protease, SULFATE ION | | Authors: | Lockbaum, G.J, Schiffer, C.A. | | Deposit date: | 2018-05-18 | | Release date: | 2018-12-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.908 Å) | | Cite: | Structural Adaptation of Darunavir Analogues against Primary Mutations in HIV-1 Protease.
ACS Infect Dis, 5, 2019
|
|
4WWN
 
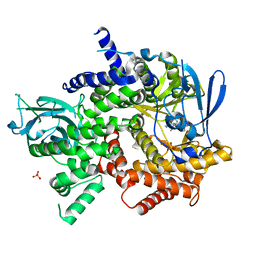 | | Crystal structure of human PI3K-gamma in complex with (S)-N-(1-(7-fluoro-2-(pyridin-2-yl)quinolin-3-yl)ethyl)-9H-purin-6-amine AMG319 inhibitor | | Descriptor: | N-{(1S)-1-[7-fluoro-2-(pyridin-2-yl)quinolin-3-yl]ethyl}-9H-purin-6-amine, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION | | Authors: | Whittington, D.A, Tang, J, Yakowec, P. | | Deposit date: | 2014-11-11 | | Release date: | 2014-12-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery and in Vivo Evaluation of (S)-N-(1-(7-Fluoro-2-(pyridin-2-yl)quinolin-3-yl)ethyl)-9H-purin-6-amine (AMG319) and Related PI3K delta Inhibitors for Inflammation and Autoimmune Disease.
J.Med.Chem., 58, 2015
|
|
4WUN
 
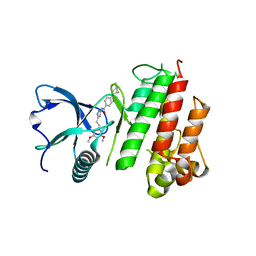 | | Structure of FGFR1 in complex with AZD4547 (N-{3-[2-(3,5-DIMETHOXYPHENYL)ETHYL]-1H-PYRAZOL-5-YL}-4-[(3R,5S)-3,5-DIMETHYLPIPERAZIN-1-YL]BENZAMIDE) at 1.65 angstrom | | Descriptor: | Fibroblast growth factor receptor 1, N-{3-[2-(3,5-dimethoxyphenyl)ethyl]-1H-pyrazol-5-yl}-4-[(3R,5S)-3,5-dimethylpiperazin-1-yl]benzamide | | Authors: | Squire, C.J, Yosaatmadja, C.J. | | Deposit date: | 2014-11-02 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The 1.65 angstrom resolution structure of the complex of AZD4547 with the kinase domain of FGFR1 displays exquisite molecular recognition.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
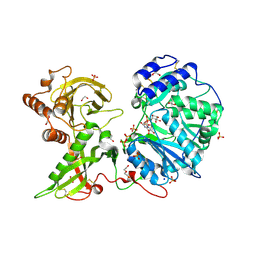
6EJ7
 
 | |
