7F4B
 
 | | The crystal structure of the immature apo-enzyme of homoserine dehydrogenase from the hyperthermophilic archaeon Sulfurisphaera tokodaii. | | Descriptor: | MAGNESIUM ION, homoserine dehydrogenase | | Authors: | Kurihara, E, Kubota, T, Watanabe, K, Ogata, K, Kaneko, R, Oshima, T, Yoshimune, K, Goto, M. | | Deposit date: | 2021-06-18 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Conformational changes in the catalytic region are responsible for heat-induced activation of hyperthermophilic homoserine dehydrogenase.
Commun Biol, 5, 2022
|
|
4RP4
 
 | |
6O3P
 
 | |
4RP3
 
 | | Crystal Structure of the L27 Domain of Discs Large 1 (target ID NYSGRC-010766) from Drosophila melanogaster bound to a potassium ion (space group P212121) | | Descriptor: | CHLORIDE ION, Disks large 1 tumor suppressor protein, FORMIC ACID, ... | | Authors: | Ghosh, A, Ramagopal, U, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-10-29 | | Release date: | 2014-11-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Structures of the L27 Domain of Disc Large Homologue 1 Protein Illustrate a Self-Assembly Module.
Biochemistry, 57, 2018
|
|
7KKJ
 
 | | Structure of anti-SARS-CoV-2 Spike nanobody mNb6 | | Descriptor: | CHLORIDE ION, SULFATE ION, Synthetic nanobody mNb6 | | Authors: | Schoof, M.S, Faust, B.F, Saunders, R.A, Sangwan, S, Rezelj, V, Hoppe, N, Boone, M, Billesboelle, C.B, Puchades, C, Azumaya, C.M, Kratochvil, H.T, Zimanyi, M, Desphande, I, Liang, J, Dickinson, S, Nguyen, H.C, Chio, C.M, Merz, G.E, Thompson, M.C, Diwanji, D, Schaefer, K, Anand, A.A, Dobzinski, N, Zha, B.S, Simoneau, C.R, Leon, K, White, K.M, Chio, U.S, Gupta, M, Jin, M, Li, F, Liu, Y, Zhang, K, Bulkley, D, Sun, M, Smith, A.M, Rizo, A.N, Moss, F, Brilot, A.F, Pourmal, S, Trenker, R, Pospiech, T, Gupta, S, Barsi-Rhyne, B, Belyy, V, Barile-Hill, A.W, Nock, S, Liu, Y, Krogan, N.J, Ralston, C.Y, Swaney, D.L, Garcia-Sastre, A, Ott, M, Vignuzzi, M, Walter, P, Manglik, A, QCRG Structural Biology Consortium | | Deposit date: | 2020-10-27 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | An ultrapotent synthetic nanobody neutralizes SARS-CoV-2 by stabilizing inactive Spike.
Science, 370, 2020
|
|
6H19
 
 | | Crystal structure of ethyl-paraoxon inhibited recombinant human bile salt activated lipase (aged form) | | Descriptor: | ACETATE ION, Bile salt-activated lipase, SODIUM ION, ... | | Authors: | Touvrey, C, Brazzolotto, X, Nachon, F. | | Deposit date: | 2018-07-11 | | Release date: | 2019-03-27 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | X-ray structures of human bile-salt activated lipase conjugated to nerve agents surrogates.
Toxicology, 411, 2019
|
|
4WXJ
 
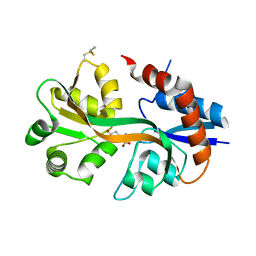 | | Drosophila muscle GluRIIB complex with glutamate | | Descriptor: | GLUTAMIC ACID, Glutamate receptor IIB,Glutamate receptor IIB | | Authors: | Dharkar, P, Mayer, M.L. | | Deposit date: | 2014-11-13 | | Release date: | 2015-04-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Functional reconstitution of Drosophila melanogaster NMJ glutamate receptors.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
6H0T
 
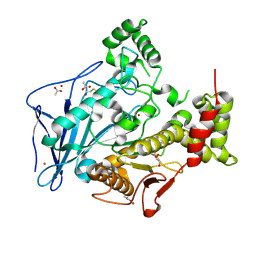 | | Crystal structure of native recombinant human bile salt activated lipase | | Descriptor: | ACETATE ION, Bile salt-activated lipase, GLYCEROL, ... | | Authors: | Touvrey, C, Brazzolotto, X, Nachon, F. | | Deposit date: | 2018-07-10 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray structures of human bile-salt activated lipase conjugated to nerve agents surrogates.
Toxicology, 411, 2019
|
|
7KKK
 
 | | SARS-CoV-2 Spike in complex with neutralizing nanobody Nb6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Schoof, M.S, Faust, B.F, Saunders, R.A, Sangwan, S, Rezelj, V, Hoppe, N, Boone, M, Billesboelle, C.B, Puchades, C, Azumaya, C.M, Kratochvil, H.T, Zimanyi, M, Desphande, I, Liang, J, Dickinson, S, Nguyen, H.C, Chio, C.M, Merz, G.E, Thompson, M.C, Diwanji, D, Schaefer, K, Anand, A.A, Dobzinski, N, Zha, B.S, Simoneau, C.R, Leon, K, White, K.M, Chio, U.S, Gupta, M, Jin, M, Li, F, Liu, Y, Zhang, K, Bulkley, D, Sun, M, Smith, A.M, Rizo, A.N, Moss, F, Brilot, A.F, Pourmal, S, Trenker, R, Pospiech, T, Gupta, S, Barsi-Rhyne, B, Belyy, V, Barile-Hill, A.W, Nock, S, Liu, Y, Krogan, N.J, Ralston, C.Y, Swaney, D.L, Garcia-Sastre, A, Ott, M, Vignuzzi, M, Walter, P, Manglik, A, QCRG Structural Biology Consortium | | Deposit date: | 2020-10-27 | | Release date: | 2020-11-11 | | Last modified: | 2021-04-21 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | An ultrapotent synthetic nanobody neutralizes SARS-CoV-2 by stabilizing inactive Spike.
Science, 370, 2020
|
|
4RP5
 
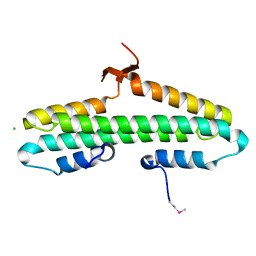 | |
7KKL
 
 | | SARS-CoV-2 Spike in complex with neutralizing nanobody mNb6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Schoof, M.S, Faust, B.F, Saunders, R.A, Sangwan, S, Rezelj, V, Hoppe, N, Boone, M, Billesboelle, C.B, Puchades, C, Azumaya, C.M, Kratochvil, H.T, Zimanyi, M, Desphande, I, Liang, J, Dickinson, S, Nguyen, H.C, Chio, C.M, Merz, G.E, Thompson, M.C, Diwanji, D, Schaefer, K, Anand, A.A, Dobzinski, N, Zha, B.S, Simoneau, C.R, Leon, K, White, K.M, Chio, U.S, Gupta, M, Jin, M, Li, F, Liu, Y, Zhang, K, Bulkley, D, Sun, M, Smith, A.M, Rizo, A.N, Moss, F, Brilot, A.F, Pourmal, S, Trenker, R, Pospiech, T, Gupta, S, Barsi-Rhyne, B, Belyy, V, Barile-Hill, A.W, Nock, S, Liu, Y, Krogan, N.J, Ralston, C.Y, Swaney, D.L, Garcia-Sastre, A, Ott, M, Vignuzzi, M, Walter, P, Manglik, A, QCRG Structural Biology Consortium | | Deposit date: | 2020-10-27 | | Release date: | 2020-11-11 | | Last modified: | 2021-04-21 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | An ultrapotent synthetic nanobody neutralizes SARS-CoV-2 by stabilizing inactive Spike.
Science, 370, 2020
|
|
7JLU
 
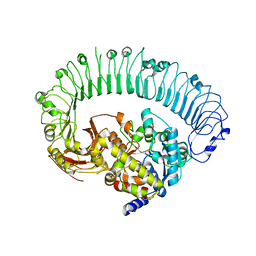 | | Structure of the activated Roq1 resistosome directly recognizing the pathogen effector XopQ | | Descriptor: | CALCIUM ION, Disease resistance protein Roq1, XopQ | | Authors: | Martin, R, Qi, T, Zhang, H, Lui, F, King, M, Toth, C, Nogales, E, Staskawicz, B.J. | | Deposit date: | 2020-07-30 | | Release date: | 2020-12-02 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of the activated ROQ1 resistosome directly recognizing the pathogen effector XopQ.
Science, 370, 2020
|
|
7JLX
 
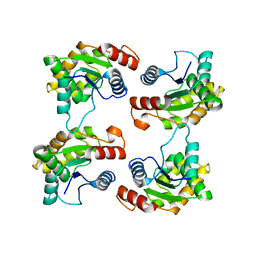 | | Structure of the activated Roq1 resistosome directly recognizing the pathogen effector XopQ (TIR domains) | | Descriptor: | Disease resistance protein Roq1 | | Authors: | Martin, R, Qi, T, Zhang, H, Lui, F, King, M, Toth, C, Nogales, E, Staskawicz, B.J. | | Deposit date: | 2020-07-30 | | Release date: | 2020-12-02 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structure of the activated ROQ1 resistosome directly recognizing the pathogen effector XopQ.
Science, 370, 2020
|
|
7KTR
 
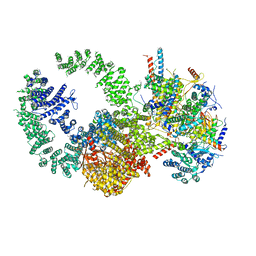 | | Cryo-EM structure of the human SAGA coactivator complex (TRRAP, core) | | Descriptor: | Ataxin-7, INOSITOL HEXAKISPHOSPHATE, Isoform 3 of Transcription factor SPT20 homolog, ... | | Authors: | Herbst, D.A, Esbin, M.N, Nogales, E. | | Deposit date: | 2020-11-24 | | Release date: | 2021-11-10 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Structure of the human SAGA coactivator complex.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7KTS
 
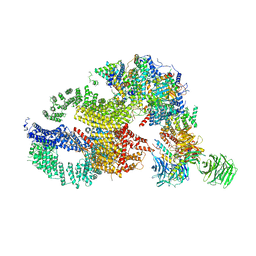 | | Negative stain EM structure of the human SAGA coactivator complex (TRRAP, core, splicing module) | | Descriptor: | Ataxin-7, Isoform 3 of Transcription factor SPT20 homolog, STAGA complex 65 subunit gamma, ... | | Authors: | Herbst, D.A, Esbin, M.N, Nogales, E. | | Deposit date: | 2020-11-24 | | Release date: | 2021-11-10 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (19.09 Å) | | Cite: | Structure of the human SAGA coactivator complex.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6ZEA
 
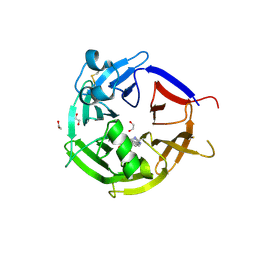 | | Strictosidine Synthase from Catharanthus roseus in complex with racemic 1-methyl-2,3,4,9-tetrahydro-1H-beta-carboline | | Descriptor: | (1~{R})-1-methyl-2,3,4,9-tetrahydro-1~{H}-pyrido[3,4-b]indole, (1~{S})-1-methyl-2,3,4,9-tetrahydro-1~{H}-pyrido[3,4-b]indole, 1,2-ETHANEDIOL, ... | | Authors: | Eger, E, Sharma, M, Kroutil, W, Grogan, G. | | Deposit date: | 2020-06-16 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Strictosidine Synthase from Catharanthus roseus in complex with racemic 1-methyl-2,3,4,9-tetrahydro-1H-beta-carboline
To Be Published
|
|
4E08
 
 | |
7OSN
 
 | | IRED361 from Micromonospora sp. in complex with NADP+ | | Descriptor: | 6-phosphogluconate dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Gilio, A.K, Harawa, V, Turner, N, Grogan, G.J. | | Deposit date: | 2021-06-09 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Synthesis of Stereoenriched Piperidines via Chemo-Enzymatic Dearomatization of Activated Pyridines.
J.Am.Chem.Soc., 144, 2022
|
|
5KDD
 
 | | Apo-structure of humanised RadA-mutant humRadA22 | | Descriptor: | DNA repair and recombination protein RadA, SULFATE ION | | Authors: | Fischer, G, Marsh, M, Moschetti, T, Sharpe, T, Scott, D, Morgan, M, Ng, H, Skidmore, J, Venkitaraman, A, Abell, C, Blundell, T.L, Hyvonen, M. | | Deposit date: | 2016-06-08 | | Release date: | 2016-10-19 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Engineering Archeal Surrogate Systems for the Development of Protein-Protein Interaction Inhibitors against Human RAD51.
J.Mol.Biol., 428, 2016
|
|
7RYA
 
 | | S. CEREVISIAE CYP51 I471T MUTANT COMPLEXED WITH ITRACONAZOLE | | Descriptor: | 2-[(2R)-butan-2-yl]-4-{4-[4-(4-{[(2R,4S)-2-(2,4-dichlorophenyl)-2-(1H-1,2,4-triazol-1-ylmethyl)-1,3-dioxolan-4-yl]methoxy}phenyl)piperazin-1-yl]phenyl}-2,4-dihydro-3H-1,2,4-triazol-3-one, Lanosterol 14-alpha demethylase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Graham, D.O, Wilson, R.K, Ruma, Y.N, Keniya, M.V, Tyndall, J.D, Monk, B.C. | | Deposit date: | 2021-08-24 | | Release date: | 2021-09-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Insights into the Azole Resistance of the Candida albicans Darlington Strain Using Saccharomyces cerevisiae Lanosterol 14 alpha-Demethylase as a Surrogate.
J Fungi (Basel), 7, 2021
|
|
7RY9
 
 | | S. CEREVISIAE CYP51 I471T mutant COMPLEXED WITH Voriconazole | | Descriptor: | Lanosterol 14-alpha demethylase, PROTOPORPHYRIN IX CONTAINING FE, TETRAETHYLENE GLYCOL, ... | | Authors: | Graham, D.O, Wilson, R.K, Ruma, Y.N, Keniya, M.V, Tyndall, J.D, Monk, B.C. | | Deposit date: | 2021-08-24 | | Release date: | 2021-09-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Insights into the Azole Resistance of the Candida albicans Darlington Strain Using Saccharomyces cerevisiae Lanosterol 14 alpha-Demethylase as a Surrogate.
J Fungi (Basel), 7, 2021
|
|
7RY8
 
 | | S. CEREVISIAE CYP51 Y140H mutant COMPLEXED WITH Voriconazole | | Descriptor: | Lanosterol 14-alpha demethylase, PENTAETHYLENE GLYCOL, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Graham, D.O, Wilson, R.K, Ruma, Y.N, Keniya, M.V, Tyndall, J.D, Monk, B.C. | | Deposit date: | 2021-08-24 | | Release date: | 2021-09-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural Insights into the Azole Resistance of the Candida albicans Darlington Strain Using Saccharomyces cerevisiae Lanosterol 14 alpha-Demethylase as a Surrogate.
J Fungi (Basel), 7, 2021
|
|
7ZBO
 
 | | Amine Dehydrogenase MATOUAmDH2 in complex with NADP+ | | Descriptor: | Amine Dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Bennett, M, Ducrot, L, Vergne-Vaxelaire, C, Grogan, G. | | Deposit date: | 2022-03-24 | | Release date: | 2022-04-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structure and Mutation of the Native Amine Dehydrogenase MATOUAmDH2.
Chembiochem, 23, 2022
|
|
7RYB
 
 | | S. CEREVISIAE CYP51 Y140H/I471T - double mutant COMPLEXED WITH Voriconazole | | Descriptor: | Lanosterol 14-alpha demethylase, PROTOPORPHYRIN IX CONTAINING FE, Voriconazole | | Authors: | Graham, D.O, Wilson, R.K, Ruma, Y.N, Keniya, M.V, Tyndall, J.D, Monk, B.C. | | Deposit date: | 2021-08-24 | | Release date: | 2021-09-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Insights into the Azole Resistance of the Candida albicans Darlington Strain Using Saccharomyces cerevisiae Lanosterol 14 alpha-Demethylase as a Surrogate.
J Fungi (Basel), 7, 2021
|
|
6EON
 
 | | Galactanase BT0290 | | Descriptor: | Beta-galactosidase, CALCIUM ION, alpha-D-galactopyranose | | Authors: | Basle, A, Munoz, J, Gilbert, H. | | Deposit date: | 2017-10-10 | | Release date: | 2017-11-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A surface endogalactanase in Bacteroides thetaiotaomicron confers keystone status for arabinogalactan degradation.
Nat Microbiol, 3, 2018
|
|
