8V87
 
 | | 60S ribosome biogenesis intermediate (Dbp10 post-catalytic structure - Overall map) | | Descriptor: | 25S rRNA (cytosine(2870)-C(5))-methyltransferase, 25S ribosomal RNA, 27S pre-rRNA (guanosine(2922)-2'-O)-methyltransferase, ... | | Authors: | Cruz, V.E, Weirich, C.S, Peddada, N, Erzberger, J.P. | | Deposit date: | 2023-12-04 | | Release date: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | The DEAD-box ATPase Dbp10/DDX54 initiates peptidyl transferase center formation during 60S ribosome biogenesis.
Nat Commun, 15, 2024
|
|
8RT5
 
 | |
8RT6
 
 | |
8RT8
 
 | |
8RT4
 
 | |
4U8T
 
 | |
2GKN
 
 | |
2GLN
 
 | |
4RB1
 
 | | Crystal structure of Magnetospirillum gryphiswaldense MSR-1 Fur-Mn2+-E. coli Fur box | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*TP*AP*AP*TP*GP*AP*TP*AP*AP*TP*CP*AP*TP*TP*AP*TP*CP*CP*GP*C)-3'), DNA-binding transcriptional dual regulator of siderophore biosynthesis and transport(Fur family), MANGANESE (II) ION | | Authors: | Deng, Z, Chen, Z. | | Deposit date: | 2014-09-12 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Mechanistic insights into metal ion activation and operator recognition by the ferric uptake regulator.
Nat Commun, 6
|
|
5RB1
 
 | | PanDDA analysis group deposition -- Crystal Structure of JMJD1B in complex with FM001700a | | Descriptor: | CHLORIDE ION, Lysine-specific demethylase 3B, MANGANESE (II) ION, ... | | Authors: | Snee, M, Nowak, R, Johansson, C, Burgess-Brown, N.A, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Oppermann, U. | | Deposit date: | 2020-03-16 | | Release date: | 2020-04-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | PanDDA analysis group deposition of Human JMJD1B screened against the DSPL Fragment Library
To Be Published
|
|
6RB1
 
 | | Human protein kinase CK2 alpha in complex with 2-cyano-2-propenamide compound 1 | | Descriptor: | (~{E})-2-cyano-3-(3-methoxy-4-oxidanyl-phenyl)-~{N}-[5-(trifluoromethyl)-1,3,4-thiadiazol-2-yl]prop-2-enamide, 1,2-ETHANEDIOL, Casein kinase II subunit alpha, ... | | Authors: | Battistutta, R, Lolli, G. | | Deposit date: | 2019-04-08 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A novel class of selective CK2 inhibitors targeting its open hinge conformation.
Eur.J.Med.Chem., 195, 2020
|
|
7RB1
 
 | | Isocitrate Lyase-1 from Mycobacterium tuberculosis covalently modified by 5-descarboxy-5-nitro-D-isocitric acid | | Descriptor: | (3E)-3-(hydroxyimino)propanoic acid, GLYCEROL, GLYOXYLIC ACID, ... | | Authors: | Krieger, I.V, Mellott, D, Meek, T, Sacchettini, J.C. | | Deposit date: | 2021-07-05 | | Release date: | 2021-12-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanism-Based Inactivation of Mycobacterium tuberculosis Isocitrate Lyase 1 by (2 R ,3 S )-2-Hydroxy-3-(nitromethyl)succinic acid.
J.Am.Chem.Soc., 143, 2021
|
|
2RB1
 
 | |
6RCB
 
 | |
6GMA
 
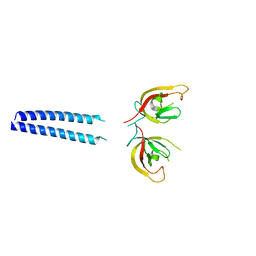 | |
6DCE
 
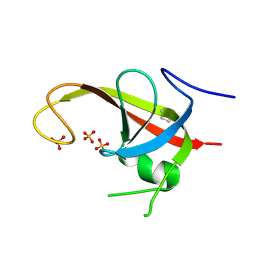 | | X-ray structure of FIP200 claw domain | | Descriptor: | RB1-inducible coiled-coil protein 1, SULFATE ION | | Authors: | Su, M.-Y, Hurley, J.H. | | Deposit date: | 2018-05-05 | | Release date: | 2019-03-06 | | Last modified: | 2019-05-01 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | FIP200 Claw Domain Binding to p62 Promotes Autophagosome Formation at Ubiquitin Condensates.
Mol. Cell, 74, 2019
|
|
4CRI
 
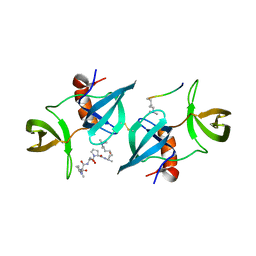 | | Crystal Structure of 53BP1 tandem tudor domains in complex with methylated K810 Rb peptide | | Descriptor: | RB1 PROTEIN, TUMOR SUPPRESSOR P53-BINDING PROTEIN 1 | | Authors: | Krojer, T, Johansson, C, Gileadi, C, Fedorov, O, Carr, S, La Thangue, N.B, Vollmar, M, Crawley, L, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Oppermann, U. | | Deposit date: | 2014-02-26 | | Release date: | 2014-08-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Lysine Methylation-Dependent Binding of 53BP1 to the Prb Tumor Suppressor.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
7SRS
 
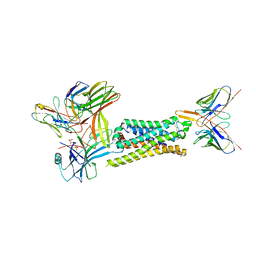 | | 5-HT2B receptor bound to LSD in complex with beta-arrestin1 obtained by cryo-electron microscopy (cryoEM) | | Descriptor: | (8alpha)-N,N-diethyl-6-methyl-9,10-didehydroergoline-8-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-hydroxytryptamine receptor 2B, ... | | Authors: | Barros-Alvarez, X, Cao, C, Panova, O, Roth, B.L, Skiniotis, G. | | Deposit date: | 2021-11-08 | | Release date: | 2022-09-21 | | Last modified: | 2022-10-19 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Signaling snapshots of a serotonin receptor activated by the prototypical psychedelic LSD.
Neuron, 110, 2022
|
|
6GYB
 
 | | Cryo-EM structure of the bacteria-killing type IV secretion system core complex from Xanthomonas citri | | Descriptor: | VirB10 protein, VirB7, VirB9 protein | | Authors: | Sgro, G.G, Costa, T.R.D, Farah, C.S, Waksman, G. | | Deposit date: | 2018-06-28 | | Release date: | 2018-10-24 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Cryo-EM structure of the bacteria-killing type IV secretion system core complex from Xanthomonas citri.
Nat Microbiol, 3, 2018
|
|
8GO8
 
 | | Structure of beta-arrestin1 in complex with a phosphopeptide corresponding to the human C5a anaphylatoxin chemotactic receptor 1, C5aR1 | | Descriptor: | Beta-arrestin-1, C5a anaphylatoxin chemotactic receptor 1, Fab30 heavy chain, ... | | Authors: | Maharana, J, Sarma, P, Yadav, M.K, Banerjee, R, Shukla, A.K. | | Deposit date: | 2022-08-24 | | Release date: | 2023-05-17 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Structural snapshots uncover a key phosphorylation motif in GPCRs driving beta-arrestin activation.
Mol.Cell, 83, 2023
|
|
8GP3
 
 | | Structure of beta-arrestin1 in complex with a phosphopeptide corresponding to the human C-X-C chemokine receptor type 4, CXCR4 | | Descriptor: | Beta-arrestin-1, C-X-C chemokine receptor type 4, Fab30 Heavy Chain, ... | | Authors: | Maharana, J, Sarma, P, Yadav, M.K, Banerjee, R, Shukla, A.K. | | Deposit date: | 2022-08-25 | | Release date: | 2023-05-17 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structural snapshots uncover a key phosphorylation motif in GPCRs driving beta-arrestin activation.
Mol.Cell, 83, 2023
|
|
6TKO
 
 | |
8HST
 
 | |
3GC3
 
 | | Crystal Structure of Arrestin2S and Clathrin | | Descriptor: | Beta-arrestin-1, Clathrin heavy chain 1 | | Authors: | Williams, J.C, Kang, D.S. | | Deposit date: | 2009-02-21 | | Release date: | 2009-08-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of an arrestin2-clathrin complex reveals a novel clathrin binding domain that modulates receptor trafficking.
J.Biol.Chem., 284, 2009
|
|
3GD1
 
 | | Structure of an Arrestin/Clathrin complex reveals a novel clathrin binding domain that modulates receptor trafficking | | Descriptor: | Beta-arrestin-1, Clathrin heavy chain 1, clathrin | | Authors: | Kang, D.S, Kern, R.C, Puthenveedu, M.A, von Zastrow, M, Williams, J.C, Benovic, J.L. | | Deposit date: | 2009-02-23 | | Release date: | 2009-08-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of an arrestin2-clathrin complex reveals a novel clathrin binding domain that modulates receptor trafficking
J.Biol.Chem., 284, 2009
|
|
