5Z30
 
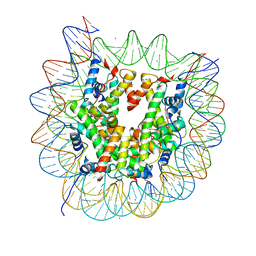 | | The crystal structure of the nucleosome containing a cancer-associated histone H2A.Z R80C mutant | | Descriptor: | CHLORIDE ION, DNA (146-MER), Histone H2A.Z, ... | | Authors: | Horikoshi, N, Arimura, Y, Kurumizaka, H. | | Deposit date: | 2018-01-05 | | Release date: | 2018-07-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Cancer-associated mutations of histones H2B, H3.1 and H2A.Z.1 affect the structure and stability of the nucleosome.
Nucleic Acids Res., 46, 2018
|
|
6QXZ
 
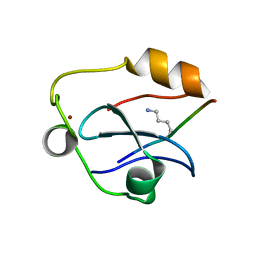 | | Solution structure of the ASHH2 CW domain with the N-terminal histone H3 tail mimicking peptide monomethylated on lysine 4 | | Descriptor: | ALA-ARG-THR-MLZ-GLN-THR-ALA-ARG-TYR, Histone-lysine N-methyltransferase ASHH2, ZINC ION | | Authors: | Dobrovolska, O, Madeleine, N, Teigen, K, Halskau, O, Bril'kov, M. | | Deposit date: | 2019-03-08 | | Release date: | 2019-12-04 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | The Arabidopsis (ASHH2) CW domain binds monomethylated K4 of the histone H3 tail through conformational selection.
Febs J., 287, 2020
|
|
7UQJ
 
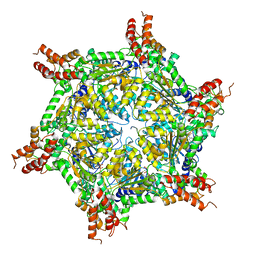 | | Cryo-EM structure of the S. cerevisiae chromatin remodeler Yta7 hexamer bound to ATPgS and histone H3 tail in state II | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATPase histone chaperone YTA7, Histone H3, ... | | Authors: | Wang, F, Feng, X, Li, H. | | Deposit date: | 2022-04-19 | | Release date: | 2023-02-01 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | The Saccharomyces cerevisiae Yta7 ATPase hexamer contains a unique bromodomain tier that functions in nucleosome disassembly.
J.Biol.Chem., 299, 2022
|
|
5C3I
 
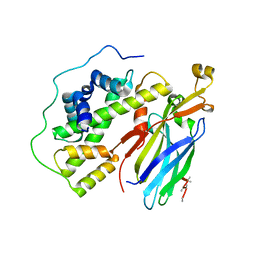 | | Crystal structure of the quaternary complex of histone H3-H4 heterodimer with chaperone ASF1 and the replicative helicase subunit MCM2 | | Descriptor: | DNA replication licensing factor MCM2,MCM2, Histone H3.1, Histone H4, ... | | Authors: | Wang, H, Wang, M, Yang, N, Xu, R.M. | | Deposit date: | 2015-06-17 | | Release date: | 2015-07-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of the quaternary complex of histone H3-H4 heterodimer with chaperone ASF1 and the replicative helicase subunit MCM2
Protein Cell, 6, 2015
|
|
3ASK
 
 | | Structure of UHRF1 in complex with histone tail | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, Histone H3.3, ZINC ION | | Authors: | Arita, K, Sugita, K, Unoki, M, Hamamoto, R, Sekiyama, N, Tochio, H, Ariyoshi, M, Shirakawa, M. | | Deposit date: | 2010-12-16 | | Release date: | 2012-01-25 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.904 Å) | | Cite: | Recognition of modification status on a histone H3 tail by linked histone reader modules of the epigenetic regulator UHRF1
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3VN5
 
 | | Crystal structure of Aquifex aeolicus RNase H3 | | Descriptor: | Ribonuclease HIII | | Authors: | Jongruja, N, You, D.J, Eiko, K, Angkawidjaja, C, Koga, Y, Takano, K, Kanaya, S. | | Deposit date: | 2011-12-22 | | Release date: | 2012-12-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure and characterization of RNase H3 from Aquifex aeolicus
Febs J., 279, 2012
|
|
1X3B
 
 | | Solution structure of the FAS1 domain of human transforming growth factor-beta induced protein IG-H3 | | Descriptor: | Transforming growth factor-beta induced protein IG-H3 | | Authors: | Yoneyama, M, Tomizawa, T, Tochio, N, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-02 | | Release date: | 2005-11-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the FAS1 domain of human transforming growth factor-beta induced protein IG-H3
To be Published
|
|
1HVV
 
 | |
3ASL
 
 | | Structure of UHRF1 in complex with histone tail | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase UHRF1, Histone H3.3, ... | | Authors: | Arita, K, Sugita, K, Unoki, M, Hamamoto, R, Sekiyama, N, Tochio, H, Ariyoshi, M, Shirakawa, M. | | Deposit date: | 2010-12-16 | | Release date: | 2012-01-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Recognition of modification status on a histone H3 tail by linked histone reader modules of the epigenetic regulator UHRF1
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
8FJM
 
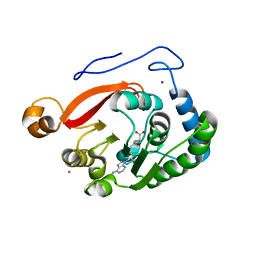 | | Crystal Structure of the Trypanosoma brucei DOT1A histone H3K76 methyltransferase in complex with AdoHcy - P212121 space group | | Descriptor: | ACETATE ION, CALCIUM ION, Histone-lysine N-methyltransferase, ... | | Authors: | Frisbie, V.S, Hashimoto, H, Debler, E.W. | | Deposit date: | 2022-12-20 | | Release date: | 2024-02-07 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Two DOT1 enzymes cooperatively mediate efficient ubiquitin-independent histone H3 lysine 76 tri-methylation in kinetoplastids.
Nat Commun, 15, 2024
|
|
2ZFN
 
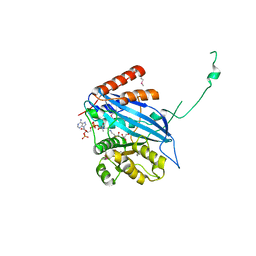 | |
2YBA
 
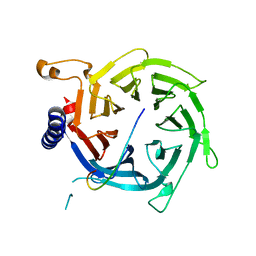 | | Crystal structure of Nurf55 in complex with histone H3 | | Descriptor: | HISTONE H3, PROBABLE HISTONE-BINDING PROTEIN CAF1 | | Authors: | Schmitges, F.W, Prusty, A.B, Faty, M, Stutzer, A, Lingaraju, G.M, Aiwazian, J, Sack, R, Hess, D, Li, L, Zhou, S, Bunker, R.D, Wirth, U, Bouwmeester, T, Bauer, A, Ly-Hartig, N, Zhao, K, Chan, H, Gu, J, Gut, H, Fischle, W, Muller, J, Thoma, N.H. | | Deposit date: | 2011-03-02 | | Release date: | 2011-05-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Histone Methylation by Prc2 is Inhibited by Active Chromatin Marks
Mol.Cell, 42, 2011
|
|
9IMZ
 
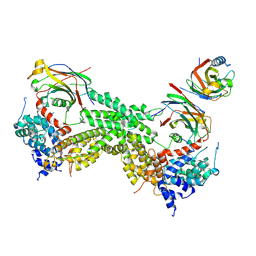 | | CODANIN-1 sequesters ASF1 by using a histone H3 mimic helix to regulate histone supply | | Descriptor: | Codanin-1, Histone chaperone ASF1A | | Authors: | Jeong, T.K, Frater, R.C.M, Yoon, J, Groth, A, Song, J.J. | | Deposit date: | 2024-07-05 | | Release date: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | CODANIN-1 sequesters ASF1 by using a histone H3 mimic helix to regulate the histone supply.
Nat Commun, 16, 2025
|
|
2LGK
 
 | | NMR Structure of UHRF1 PHD domains in a complex with histone H3 peptide | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, ZINC ION, histone H3 peptide | | Authors: | Wang, C, Shen, J, Yang, Z, Chen, P, Zhao, B, Hu, W, Lan, W, Tong, X, Wu, H, Li, G, Cao, C. | | Deposit date: | 2011-07-28 | | Release date: | 2011-09-28 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural basis for site-specific reading of unmodified R2 of histone H3 tail by UHRF1 PHD finger.
Cell Res., 21, 2011
|
|
4H9R
 
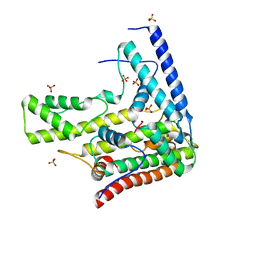 | | Complex structure 5 of DAXX(E225A)/H3.3(sub5,G90A)/H4 | | Descriptor: | Death domain-associated protein 6, Histone H3.3, Histone H4, ... | | Authors: | Elsasser, S.J, Huang, H, Lewis, P.W, Chin, J.W, Allis, D.C, Patel, D.J. | | Deposit date: | 2012-09-24 | | Release date: | 2012-10-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.197 Å) | | Cite: | DAXX chaperone envelops an H3.3/H4 dimer dictating H3.3-specific read out
To be Published
|
|
4H9O
 
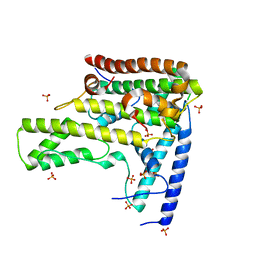 | | Complex structure 2 of DAXX/H3.3(sub5,G90M)/H4 | | Descriptor: | Death domain-associated protein 6, Histone H3.3, Histone H4, ... | | Authors: | Elsasser, S.J, Huang, H, Lewis, P.W, Chin, J.W, Allis, D.C, Patel, D.J. | | Deposit date: | 2012-09-24 | | Release date: | 2012-10-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.053 Å) | | Cite: | DAXX chaperone envelops an H3.3/H4 dimer dictating H3.3-specific read out
To be Published
|
|
4H9Q
 
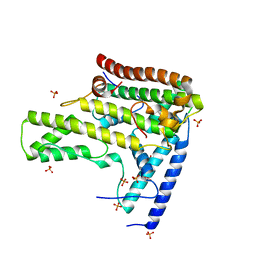 | | Complex structure 4 of DAXX(E225A)/H3.3(sub5)/H4 | | Descriptor: | Death domain-associated protein 6, Histone H3.3, Histone H4, ... | | Authors: | Elsasser, S.J, Huang, H, Lewis, P.W, Chin, J.W, Allis, D.C, Patel, D.J. | | Deposit date: | 2012-09-24 | | Release date: | 2012-10-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | DAXX chaperone envelops an H3.3/H4 dimer dictating H3.3-specific read out
To be Published
|
|
2L75
 
 | |
7CWH
 
 | | Structural basis of RACK7 PHD to read a pediatric glioblastoma-associated histone mutation H3.3G34R | | Descriptor: | Peptide from Histone H3.3, Protein kinase C-binding protein 1, ZINC ION | | Authors: | Lan, W.X, Li, Z, Jiao, F.F, Wang, C.X, Guo, R, Cao, C.Y. | | Deposit date: | 2020-08-28 | | Release date: | 2021-05-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis of RACK7 PHD domain to read a pediatric glioblastoma‐associated histone mutation H3.3G34R
Chin.J.Chem., 2021
|
|
4H9S
 
 | | Complex structure 6 of DAXX/H3.3(sub7)/H4 | | Descriptor: | Death domain-associated protein 6, Histone H3.3, Histone H4, ... | | Authors: | Elsasser, S.J, Huang, H, Lewis, P.W, Chin, J.W, Allis, D.C, Patel, D.J. | | Deposit date: | 2012-09-24 | | Release date: | 2012-10-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | DAXX chaperone envelops an H3.3/H4 dimer dictating H3.3-specific read out
To be Published
|
|
4H9P
 
 | | Complex structure 3 of DAXX/H3.3(sub5,G90A)/H4 | | Descriptor: | Death domain-associated protein 6, Histone H3.3, Histone H4, ... | | Authors: | Elsasser, S.J, Huang, H, Lewis, P.W, Chin, J.W, Allis, D.C, Patel, D.J. | | Deposit date: | 2012-09-24 | | Release date: | 2012-10-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.198 Å) | | Cite: | DAXX chaperone envelops an H3.3/H4 dimer dictating H3.3-specific read out
To be Published
|
|
4H9N
 
 | | Complex structure 1 of DAXX/H3.3(sub5)/H4 | | Descriptor: | Death domain-associated protein 6, Histone H3.3, Histone H4, ... | | Authors: | Elsasser, S.J, Huang, H, Lewis, P.W, Chin, J.W, Allis, D.C, Patel, D.J. | | Deposit date: | 2012-09-24 | | Release date: | 2012-10-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | DAXX chaperone envelops an H3.3/H4 dimer dictating H3.3-specific read out
To be Published
|
|
3AVR
 
 | | Catalytic fragment of UTX/KDM6A bound with histone H3K27me3 peptide, N-oxyalylglycine, and Ni(II) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Histone H3, ... | | Authors: | Sengoku, T, Yokoyama, S. | | Deposit date: | 2011-03-07 | | Release date: | 2011-10-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | Structural basis for histone H3 Lys 27 demethylation by UTX/KDM6A
Genes Dev., 25, 2011
|
|
8XAG
 
 | |
1Q3L
 
 | |
