4W7Y
 
 | |
4W82
 
 | | Enoyl-acyl carrier protein-reductase domain from human fatty acid synthase | | Descriptor: | CHLORIDE ION, Fatty acid synthase, MAGNESIUM ION, ... | | Authors: | Sippel, K.H, Vyas, N.K, Sankaran, B, Quiocho, F.A. | | Deposit date: | 2014-08-22 | | Release date: | 2014-10-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the human Fatty Acid synthase enoyl-acyl carrier protein-reductase domain complexed with triclosan reveals allosteric protein-protein interface inhibition.
J.Biol.Chem., 289, 2014
|
|
4W8I
 
 | | Crystal structure of LpSPL/Lpp2128, Legionella pneumophila sphingosine-1 phosphate lyase | | Descriptor: | Probable sphingosine-1-phosphate lyase | | Authors: | Stogios, P.J, Daniels, C, Skarina, T, Cuff, M, Di Leo, R, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-08-24 | | Release date: | 2014-11-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Legionella pneumophila S1P-lyase targets host sphingolipid metabolism and restrains autophagy.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4W8T
 
 | |
4W8Z
 
 | |
4W9C
 
 | | pVHL:EloB:EloC in complex with (2S,4R)-1-(3,3-dimethylbutanoyl)-4-hydroxy-N-(4-(oxazol-5-yl)benzyl)pyrrolidine-2-carboxamide (ligand 2) | | Descriptor: | (4R)-1-(3,3-dimethylbutanoyl)-4-hydroxy-N-[4-(1,3-oxazol-5-yl)benzyl]-L-prolinamide, Transcription elongation factor B polypeptide 1, Transcription elongation factor B polypeptide 2, ... | | Authors: | Gadd, M.S, Hewitt, S, Galdeano, C, van Molle, I, Ciulli, A. | | Deposit date: | 2014-08-27 | | Release date: | 2014-09-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Guided Design and Optimization of Small Molecules Targeting the Protein-Protein Interaction between the von Hippel-Lindau (VHL) E3 Ubiquitin Ligase and the Hypoxia Inducible Factor (HIF) Alpha Subunit with in Vitro Nanomolar Affinities.
J.Med.Chem., 57, 2014
|
|
4W9R
 
 | | Crystal structure of uncharacterised protein Coch_1243 from Capnocytophaga ochracea DSM 7271 | | Descriptor: | ACETATE ION, GLYCEROL, Uncharacterized protein | | Authors: | Chang, C, Wu, R, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-08-27 | | Release date: | 2014-09-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.703 Å) | | Cite: | Crystal structure of uncharacterised protein Coch_1243 from Capnocytophaga ochracea DSM 7271
To Be Published
|
|
4WA8
 
 | |
4WAJ
 
 | |
4WB0
 
 | |
4WBL
 
 | | Catalytic domain of mouse 2',3'-cyclic nucleotide 3'- phosphodiesterase, with mutation F235A | | Descriptor: | 2',3'-cyclic-nucleotide 3'-phosphodiesterase, CHLORIDE ION, GLYCEROL | | Authors: | Myllykoski, M, Raasakka, A, Kursula, P. | | Deposit date: | 2014-09-03 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Determinants of ligand binding and catalytic activity in the myelin enzyme 2',3'-cyclic nucleotide 3'-phosphodiesterase.
Sci Rep, 5, 2015
|
|
4WC4
 
 | | tRNA-processing enzyme complex 2 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Poly A polymerase, RNA (74-MER) | | Authors: | Yamashita, S, Tomita, K. | | Deposit date: | 2014-09-04 | | Release date: | 2015-04-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.501 Å) | | Cite: | Measurement of Acceptor-T Psi C Helix Length of tRNA for Terminal A76-Addition by A-Adding Enzyme.
Structure, 23, 2015
|
|
4WCF
 
 | | Trypanosoma brucei PTR1 in complex with inhibitor 9 | | Descriptor: | 3-(5-amino-1,3,4-thiadiazol-2-yl)pyridin-4-amine, ACETATE ION, GLYCEROL, ... | | Authors: | Mangani, S, Di Pisa, F, Pozzi, C. | | Deposit date: | 2014-09-04 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Exploiting the 2-Amino-1,3,4-thiadiazole Scaffold To Inhibit Trypanosoma brucei Pteridine Reductase in Support of Early-Stage Drug Discovery.
ACS Omega, 2, 2017
|
|
4WD4
 
 | | Crystal structure of human HO1 H25R | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Heme oxygenase 1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Caaveiro, J.M.M, Morante, K, Sigala, P, Tsumoto, K. | | Deposit date: | 2014-09-06 | | Release date: | 2015-09-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | In-Cell Enzymology To Probe His-Heme Ligation in Heme Oxygenase Catalysis
Biochemistry, 55, 2016
|
|
4WDU
 
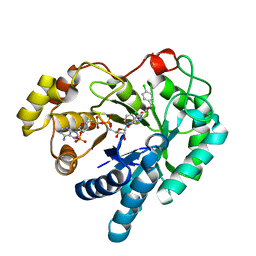 | | 17beta-HSD5 in complex with 4-chloro-N-(4-chlorobenzyl)-5-nitro-1H-pyrazole-3-carboxamide | | Descriptor: | 4-chloro-N-(4-chlorobenzyl)-5-nitro-1H-pyrazole-3-carboxamide, Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Amano, Y, Yamaguchi, T. | | Deposit date: | 2014-09-09 | | Release date: | 2015-04-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of complexes of type 5 17 beta-hydroxysteroid dehydrogenase with structurally diverse inhibitors: insights into the conformational changes upon inhibitor binding.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4V71
 
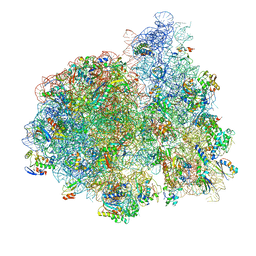 | | E. coli 70S-fMetVal-tRNAVal-tRNAfMet complex in intermediate pre-translocation state (pre2) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Blau, C, Bock, L.V, Schroder, G.F, Davydov, I, Fischer, N, Stark, H, Rodnina, M.V, Vaiana, A.C, Grubmuller, H. | | Deposit date: | 2013-10-14 | | Release date: | 2014-07-09 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (20 Å) | | Cite: | Energy barriers and driving forces in tRNA translocation through the ribosome.
Nat.Struct.Mol.Biol., 20, 2013
|
|
4WE2
 
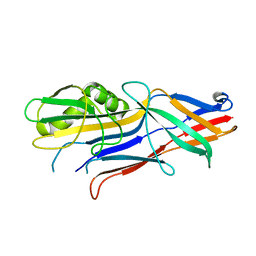 | | Donor strand complemented FaeG of F4ab fimbriae | | Descriptor: | K88 fimbrial protein AB | | Authors: | Moonens, K, Van den Broeck, I, De Kerpel, M, Deboeck, F, Raymaekers, H, Remaut, H, De Greve, H. | | Deposit date: | 2014-09-09 | | Release date: | 2015-02-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and Functional Insight into the Carbohydrate Receptor Binding of F4 Fimbriae-producing Enterotoxigenic Escherichia coli.
J.Biol.Chem., 290, 2015
|
|
4V81
 
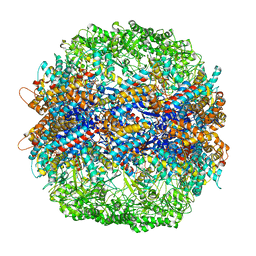 | | The crystal structure of yeast CCT reveals intrinsic asymmetry of eukaryotic cytosolic chaperonins | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, SULFATE ION, ... | | Authors: | Dekker, C, Roe, S.M, McCormack, E.A, Beuron, F, Pearl, L.H, Willison, K.R. | | Deposit date: | 2010-10-17 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | The crystal structure of yeast CCT reveals intrinsic asymmetry of eukaryotic cytosolic chaperonins.
Embo J., 30, 2011
|
|
4WEC
 
 | |
4UWX
 
 | | Structure of liprin-alpha3 in complex with mDia1 Diaphanous- inhibitory domain | | Descriptor: | LIPRIN-ALPHA-3, NICKEL (II) ION, PROTEIN DIAPHANOUS HOMOLOG 1, ... | | Authors: | Brenig, J, de Boor, S, Knyphausen, P, Kuhlmann, N, Wroblowski, S, Baldus, L, Scislowski, L, Artz, O, Trauschies, P, Baumann, U, Neundorf, I, Lammers, M. | | Deposit date: | 2014-08-15 | | Release date: | 2015-05-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural and Biochemical Basis for the Inhibitory Effect of Liprin-Alpha3 on Mouse Diaphanous 1 (Mdia1) Function.
J.Biol.Chem., 290, 2015
|
|
4URH
 
 | | High-resolution structure of partially oxidized D. fructosovorans NiFe-hydrogenase | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE3-S4 CLUSTER, GLYCEROL, ... | | Authors: | Volbeda, A, Martin, L, Barbier, E, Gutierrez-Sanz, O, DeLacey, A.L, Liebgott, P.P, Dementin, S, Rousset, M, Fontecilla-Camps, J.C. | | Deposit date: | 2014-06-30 | | Release date: | 2014-10-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Crystallographic studies of [NiFe]-hydrogenase mutants: towards consensus structures for the elusive unready oxidized states.
J. Biol. Inorg. Chem., 20, 2015
|
|
4UV3
 
 | | Structure of the curli transport lipoprotein CsgG in its membrane- bound conformation | | Descriptor: | CURLI PRODUCTION ASSEMBLY/TRANSPORT COMPONENT CSGG | | Authors: | Goyal, P, Krasteva, P.V, Gerven, N.V, Gubellini, F, Broeck, I.V.D, Troupiotis-Tsailaki, A, Jonckheere, W, Pehau-Arnaudet, G, Pinkner, J.S, Chapman, M.R, Hultgren, S.J, Howorka, S, Fronzes, R, Remaut, H. | | Deposit date: | 2014-08-04 | | Release date: | 2014-09-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.59 Å) | | Cite: | Structural and Mechanistic Insights Into the Bacterial Amyloid Secretion Channel Csgg.
Nature, 516, 2014
|
|
4V0R
 
 | | DENGUE VIRUS FULL LENGTH NS5 COMPLEXED WITH GTP AND SAH | | Descriptor: | FORMIC ACID, GLYCEROL, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Zhao, Y, Soh, S, Zheng, J, Phoo, W.W, Swaminathan, K, Cornvik, T.C, Lim, S.P, Shi, P.-Y, Lescar, J, Vasudevan, S.G, Luo, D. | | Deposit date: | 2014-09-18 | | Release date: | 2015-01-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A Crystal Structure of the Dengue Virus Ns5 Protein Reveals a Novel Inter-Domain Interface Essential for Protein Flexibility and Virus Replication.
Plos Pathog., 11, 2015
|
|
4V3Q
 
 | | Designed armadillo repeat protein with 4 internal repeats, 2nd generation C-cap and 3rd generation N-cap. | | Descriptor: | CALCIUM ION, GLYCEROL, YIII_M4_AII | | Authors: | Reichen, C, Madhurantakam, C, Pluckthun, A, Mittl, P. | | Deposit date: | 2014-10-20 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of designed armadillo-repeat proteins show propagation of inter-repeat interface effects.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
4US7
 
 | | Sulfur SAD Phased Structure of a Type IV Pilus Protein from Shewanella oneidensis | | Descriptor: | PILD PROCESSED PROTEIN, SODIUM ION, SULFATE ION | | Authors: | Gorgel, M, Boeggild, A, Ulstrup, J.J, Mueller, U, Weiss, M, Nissen, P, Boesen, T. | | Deposit date: | 2014-07-03 | | Release date: | 2015-04-29 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | High-Resolution Structure of a Type Iv Pilin from the Metal- Reducing Bacterium Shewanella Oneidensis.
Bmc Struct.Biol., 15, 2015
|
|
