6KQB
 
 | | A long chain secondary alcohol dehydrogenase of Micrococcus luteus | | Descriptor: | 3-hydroxybutyryl-CoA dehydrogenase | | Authors: | Kim, H.J, Kim, J.S. | | Deposit date: | 2019-08-16 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.261 Å) | | Cite: | Cofactor specificity engineering of a long-chain secondary alcohol dehydrogenase from Micrococcus luteus for redox-neutral biotransformation of fatty acids.
Chem.Commun.(Camb.), 55, 2019
|
|
6KQ9
 
 | | A long chain secondary alcohol dehydrogenase of Micrococcus luteus | | Descriptor: | 3-hydroxybutyryl-CoA dehydrogenase | | Authors: | Kim, H.J, Kim, J.S. | | Deposit date: | 2019-08-16 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.251 Å) | | Cite: | Cofactor specificity engineering of a long-chain secondary alcohol dehydrogenase from Micrococcus luteus for redox-neutral biotransformation of fatty acids.
Chem.Commun.(Camb.), 55, 2019
|
|
4JW2
 
 | | Selection of specific protein binders for pre-defined targets from an optimized library of artificial helicoidal repeat proteins (alphaRep) | | Descriptor: | 1,2-ETHANEDIOL, A3 artificial protein, bA3-2: binder of A3 protein | | Authors: | Guellouz, A, Valerio-Lepiniec, M, Urvoas, A, Chevrel, A, Graille, M, Fourati-Kammoun, Z, Desmadril, M, van Tilbeurgh, H, Minard, P. | | Deposit date: | 2013-03-27 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Selection of Specific Protein Binders for Pre-Defined Targets from an Optimized Library of Artificial Helicoidal Repeat Proteins (alphaRep).
Plos One, 8, 2013
|
|
1TB6
 
 | | 2.5A Crystal Structure of the Antithrombin-Thrombin-Heparin Ternary Complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2,3,4,6-tetra-O-sulfonato-alpha-D-glucopyranose-(1-4)-2,3,6-tri-O-sulfo-alpha-D-glucopyranose-(1-4)-2,3,6-tri-O-sulfonato-beta-D-glucopyranose-(1-4)-2,3-di-O-methyl-6-O-sulfonato-alpha-D-glucopyranose-(1-4)-2,3,6-tri-O-methyl-beta-D-glucopyranose-(1-4)-2,3,6-tri-O-methyl-alpha-D-glucopyranose-(1-4)-2,3,6-tri-O-methyl-beta-D-glucopyranose-(1-4)-2,3,6-tri-O-methyl-alpha-D-glucopyranose-(1-4)-2,3,6-tri-O-methyl-beta-D-glucopyranose-(1-4)-2,3,6-tri-O-methyl-alpha-D-glucopyranose-(1-4)-2,3,6-tri-O-methyl-beta-D-glucopyranose-(1-4)-2,3-di-O-methyl-6-O-sulfonato-alpha-D-glucopyranose-(1-4)-2,3-di-O-methyl-beta-D-glucopyranuronic acid-(1-4)-2,3,6-tri-O-sulfo-alpha-D-glucopyranose-(1-4)-2,3-di-O-methyl-alpha-L-idopyranuronic acid-(1-4)-methyl 3-O-methyl-2,6-di-O-sulfo-alpha-D-glucopyranoside, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Li, W, Johnson, D.J, Esmon, C.T, Huntington, J.A. | | Deposit date: | 2004-05-19 | | Release date: | 2004-08-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the antithrombin-thrombin-heparin ternary complex reveals the antithrombotic mechanism of heparin.
Nat.Struct.Mol.Biol., 11, 2004
|
|
1EJ4
 
 | | COCRYSTAL STRUCTURE OF EIF4E/4E-BP1 PEPTIDE | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, EUKARYOTIC INITIATION FACTOR 4E, EUKARYOTIC TRANSLATION INITIATION FACTOR 4E BINDING PROTEIN 1 | | Authors: | Marcotrigiano, J, Gingras, A.-C, Sonenberg, N, Burley, S.K. | | Deposit date: | 2000-02-28 | | Release date: | 2000-03-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Cap-dependent translation initiation in eukaryotes is regulated by a molecular mimic of eIF4G.
Mol.Cell, 3, 1999
|
|
1G94
 
 | | CRYSTAL STRUCTURE ANALYSIS OF THE TERNARY COMPLEX BETWEEN PSYCHROPHILIC ALPHA AMYLASE FROM PSEUDOALTEROMONAS HALOPLANCTIS IN COMPLEX WITH A HEPTA-SACCHARIDE AND A TRIS MOLECULE | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4,6-dideoxy-4-{[(1S,5R,6S)-3-formyl-5,6-dihydroxy-4-oxocyclohex-2-en-1-yl]amino}-alpha-D-xylo-hex-5-enopyranose-(1-4)-alpha-D-glucopyranose, 4,6-dideoxy-4-{[(1S,5R,6S)-3-formyl-5,6-dihydroxy-4-oxocyclohex-2-en-1-yl]amino}-alpha-D-xylo-hex-5-enopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Aghajari, N, Roth, M, Haser, R. | | Deposit date: | 2000-11-22 | | Release date: | 2001-12-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystallographic evidence of a transglycosylation reaction: ternary complexes of a psychrophilic alpha-amylase.
Biochemistry, 41, 2002
|
|
8OXN
 
 | | CRYSTAL STRUCTURE OF THE COFACTOR-DEVOID 1-H-3-HYDROXY-4- OXOQUINALDINE 2,4-DIOXYGENASE (HOD) S101A VARIANT COMPLEXED WITH 2-METHYL-QUINOLIN-4(1H)-ONE UNDER NORMOXYC CONDITIONS | | Descriptor: | 1H-3-hydroxy-4-oxoquinaldine 2,4-dioxygenase, 2-methyl-quinolin-4(1H)-one, GLYCEROL, ... | | Authors: | Bui, S, Steiner, R.A. | | Deposit date: | 2023-05-02 | | Release date: | 2024-01-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Evolutionary adaptation from hydrolytic to oxygenolytic catalysis at the alpha / beta-hydrolase fold.
Chem Sci, 14, 2023
|
|
8OXT
 
 | | CRYSTAL STRUCTURE OF THE COFACTOR-DEVOID 1-H-3-HYDROXY-4- OXOQUINALDINE 2,4-DIOXYGENASE (HOD) H251A VARIANT COMPLEXED WITH N-ACETYLANTHRANILATE AS RESULT OF IN CRYSTALLO TURNOVER OF ITS NATURAL SUBSTRATE 1-H-3-HYDROXY-4- OXOQUINALDINE UNDER HYPEROXIC CONDITIONS | | Descriptor: | 1H-3-hydroxy-4-oxoquinaldine 2,4-dioxygenase, 2-(ACETYLAMINO)BENZOIC ACID, GLYCEROL, ... | | Authors: | Bui, S, Steiner, R.A. | | Deposit date: | 2023-05-02 | | Release date: | 2024-01-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | Evolutionary adaptation from hydrolytic to oxygenolytic catalysis at the alpha / beta-hydrolase fold.
Chem Sci, 14, 2023
|
|
4Y30
 
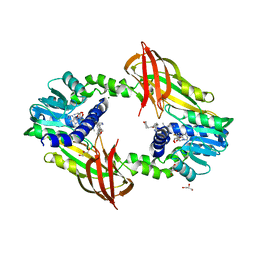 | | Crystal structure of human protein arginine methyltransferase PRMT6 bound to SAH and EPZ020411 | | Descriptor: | GLYCEROL, MAGNESIUM ION, N,N'-dimethyl-N-({3-[4-({trans-3-[2-(tetrahydro-2H-pyran-4-yl)ethoxy]cyclobutyl}oxy)phenyl]-1H-pyrazol-4-yl}methyl)ethane-1,2-diamine, ... | | Authors: | Swinger, K.K, Boriack-Sjodin, P.A. | | Deposit date: | 2015-02-10 | | Release date: | 2015-04-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Aryl Pyrazoles as Potent Inhibitors of Arginine Methyltransferases: Identification of the First PRMT6 Tool Compound.
Acs Med.Chem.Lett., 6, 2015
|
|
1MTP
 
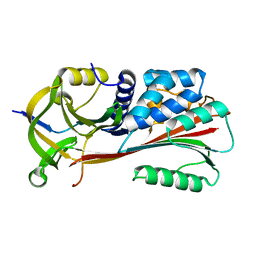 | | The X-ray crystal structure of a serpin from a thermophilic prokaryote | | Descriptor: | Serine Proteinase Inhibitor (SERPIN), Chain A, Chain B | | Authors: | Irving, J.A, Cabrita, L.D, Rossjohn, J, Pike, R.N, Bottomley, S.P, Whisstock, J.C. | | Deposit date: | 2002-09-21 | | Release date: | 2003-04-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The 1.5 A crystal structure of a prokaryote serpin: controlling conformational change in a heated environment
Structure, 11, 2003
|
|
1MVT
 
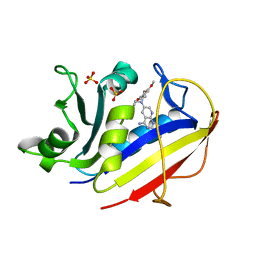 | | Analysis of Two Polymorphic Forms of a Pyrido[2,3-d]pyrimidine N9-C10 Reverse-Bridge Antifolate Binary Complex with Human Dihydrofolate Reductase | | Descriptor: | 2,4-DIAMINO-6-[N-(3',4',5'-TRIMETHOXYBENZYL)-N-METHYLAMINO]PYRIDO[2,3-D]PYRIMIDINE, Dihydrofolate Reductase, SULFATE ION | | Authors: | Cody, V, Galitsky, N, Luft, J.R, Pangborn, W.A, Gangjee, A. | | Deposit date: | 2002-09-26 | | Release date: | 2003-04-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Analysis of two polymorphic forms of a pyrido[2,3-d]pyrimidine N9-C10 reversed-bridge antifolate binary complex with human dihydrofolate reductase.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
5V2G
 
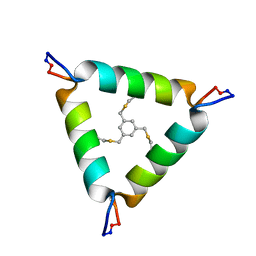 | | De Novo Design of Novel Covalent Constrained Meso-size Peptide Scaffolds with Unique Tertiary Structures | | Descriptor: | 1,3,5-tris(bromomethyl)benzene, 20-mer Peptide | | Authors: | Dang, B, Wu, H, Mulligan, V.K, Mravic, M, Wu, Y, Lemmin, T, Ford, A, Silva, D, Baker, D, DeGrado, W.F. | | Deposit date: | 2017-03-03 | | Release date: | 2017-09-27 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | De novo design of covalently constrained mesosize protein scaffolds with unique tertiary structures.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6Y4Q
 
 | | Structure of a stapled peptide bound to MDM2 | | Descriptor: | ACE-LEU-THR-PHE-GLY-GLU-TYR-TRP-ALA-GLN-LEU-ALA-SER, E3 ubiquitin-protein ligase Mdm2, ~{N}-[(1-ethyl-1,2,3-triazol-4-yl)methyl]-~{N},5-dimethyl-4-[2-[2-methyl-5-[methyl-[(1-propyl-1,2,3-triazol-4-yl)methyl]carbamoyl]thiophen-3-yl]cyclopenten-1-yl]thiophene-2-carboxamide | | Authors: | Pantelejevs, T, Bakanovych, I. | | Deposit date: | 2020-02-22 | | Release date: | 2020-05-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Diarylethene moiety as an enthalpy-entropy switch: photoisomerizable stapled peptides for modulating p53/MDM2 interaction.
Org.Biomol.Chem., 18, 2020
|
|
4RGY
 
 | |
8SUO
 
 | | BA.2/AZD1061/AZD3152 structure analysis | | Descriptor: | AZD1061 heavy chain, AZD1061 light chain, AZD3152 heavy chain, ... | | Authors: | Oganesyan, V, van Dyk, N, Dippel, A, Barnes, A, O'Connor, E. | | Deposit date: | 2023-05-12 | | Release date: | 2024-05-15 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | AZD3152 neutralizes SARS-CoV-2 historical and contemporary variants and is protective in hamsters and well tolerated in adults.
Sci Transl Med, 16, 2024
|
|
6SDZ
 
 | |
8ORO
 
 | | CRYSTAL STRUCTURE OF THE COFACTOR-DEVOID 1-H-3-HYDROXY-4- OXOQUINALDINE 2,4-DIOXYGENASE (HOD) S101A VARIANT COMPLEXED WITH 2-METHYL-QUINOLIN-4(1H)-ONE UNDER HYPEROXYC CONDITIONS | | Descriptor: | 1H-3-hydroxy-4-oxoquinaldine 2,4-dioxygenase, 2-methyl-quinolin-4(1H)-one, D(-)-TARTARIC ACID, ... | | Authors: | Bui, S, Steiner, R.A. | | Deposit date: | 2023-04-15 | | Release date: | 2024-01-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Evolutionary adaptation from hydrolytic to oxygenolytic catalysis at the alpha / beta-hydrolase fold.
Chem Sci, 14, 2023
|
|
5WOD
 
 | |
3IYI
 
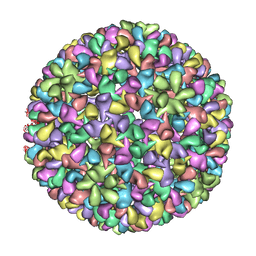 | | P22 expanded head coat protein structures reveal a novel mechanism for capsid maturation: Stability without auxiliary proteins or chemical cross-links | | Descriptor: | P22 coat protein in procapsid shells | | Authors: | Parent, K.N, Khayat, R, Tu, L.H, Suhanovsky, M.M, Cortines, J.R, Teschke, C.M, Johnson, J.E, Baker, T.S. | | Deposit date: | 2009-12-14 | | Release date: | 2010-03-31 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (9.1 Å) | | Cite: | P22 coat protein structures reveal a novel mechanism for capsid maturation: stability without auxiliary proteins or chemical crosslinks
Structure, 18, 2010
|
|
2VUZ
 
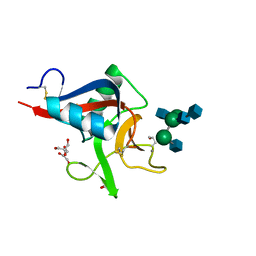 | | Crystal structure of Codakine in complex with biantennary nonasaccharide at 1.7A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CODAKINE, ... | | Authors: | Gourdine, J.P, Cioci, G.C, Miguet, L, Unverzagt, C, Varrot, A, Gauthier, C, Smith-Ravin, E.J, Imberty, A. | | Deposit date: | 2008-06-02 | | Release date: | 2008-08-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | High Affinity Interaction between a Bivalve C-Type Lectin and a Biantennary Complex-Type N-Glycan Revealed by Crystallography and Microcalorimetry.
J.Biol.Chem., 283, 2008
|
|
5WD6
 
 | | bovine salivary protein form 30b | | Descriptor: | CALCIUM ION, Short palate, lung and nasal epithelium carcinoma-associated protein 2B | | Authors: | Zhang, H, Arcus, V.L. | | Deposit date: | 2017-07-04 | | Release date: | 2018-07-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The three dimensional structure of Bovine Salivary Protein 30b (BSP30b) and its interaction with specific rumen bacteria.
Plos One, 14, 2019
|
|
3OWB
 
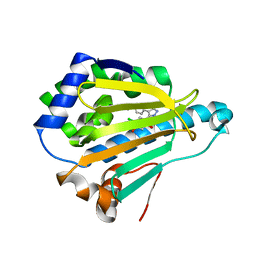 | | Crystal Structure of HSP90 with VER-49009 | | Descriptor: | 5-(5-CHLORO-2,4-DIHYDROXYPHENYL)-N-ETHYL-4-(4-METHOXYPHENYL)-1H-PYRAZOLE-3-CARBOXAMIDE, Heat shock protein HSP 90-alpha | | Authors: | Park, C.H. | | Deposit date: | 2010-09-17 | | Release date: | 2011-09-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | N-aryl-benzimidazolones as novel small molecule HSP90 inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
5V2O
 
 | | De Novo Design of Novel Covalent Constrained Meso-size Peptide Scaffolds with Unique Tertiary Structures | | Descriptor: | 1,3,5-BENZENETRICARBOXYLIC ACID, GLYCEROL, NONAETHYLENE GLYCOL, ... | | Authors: | Dang, B, Wu, H, Mulligan, V.K, Mravic, M, Wu, Y, Lemmin, T, Ford, A, Silva, D, Baker, D, DeGrado, W.F. | | Deposit date: | 2017-03-06 | | Release date: | 2017-10-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | De novo design of covalently constrained mesosize protein scaffolds with unique tertiary structures.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
3RSM
 
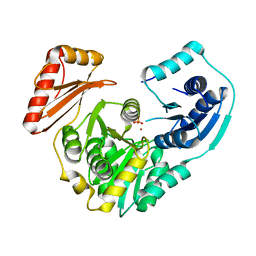 | | Crystal structure of S108C mutant of PMM/PGM | | Descriptor: | PHOSPHATE ION, Phosphomannomutase/phosphoglucomutase, ZINC ION | | Authors: | Akella, A, Anbanandam, A, Kelm, A, Wei, Y, Mehra-Chaudhary, R, Beamer, L, Van Doren, S. | | Deposit date: | 2011-05-02 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Solution NMR of a 463-residue phosphohexomutase: domain 4 mobility, substates, and phosphoryl transfer defect.
Biochemistry, 51, 2012
|
|
3RCE
 
 | | Bacterial oligosaccharyltransferase PglB | | Descriptor: | MAGNESIUM ION, Oligosaccharide transferase to N-glycosylate proteins, Substrate Mimic Peptide | | Authors: | Lizak, C, Gerber, S, Numao, S, Aebi, M, Locher, K.P. | | Deposit date: | 2011-03-31 | | Release date: | 2011-06-15 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | X-ray structure of a bacterial oligosaccharyltransferase.
Nature, 474, 2011
|
|
