8VVO
 
 | | Structure of FabS1CE2-EPR1-1 in complex with the erythropoietin receptor | | Descriptor: | CHLORIDE ION, Erythropoietin receptor, S1CE2 VARIANT OF FAB-EPR-1 heavy chain, ... | | Authors: | Singer, A.U, Bruce, H.A, Pavlenco, A, Ploder, L, Luu, G, Blazer, L, Adams, J.J, Sidhu, S.S. | | Deposit date: | 2024-01-31 | | Release date: | 2024-07-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Antigen-binding fragments with improved crystal lattice packing and enhanced conformational flexibility at the elbow region as crystallization chaperones.
Protein Sci., 33, 2024
|
|
5FFJ
 
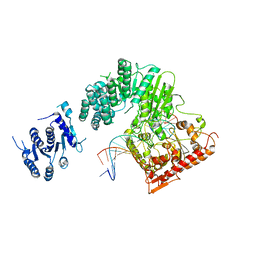 | |
6NNP
 
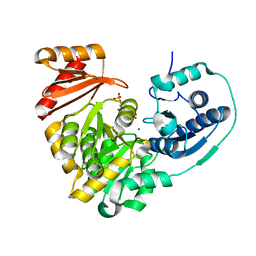 | |
7O0S
 
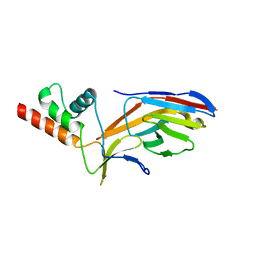 | |
8S3X
 
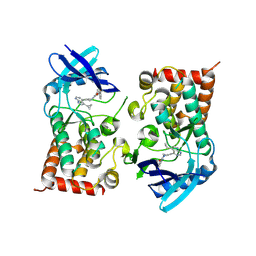 | | LIM Domain Kinase 2 (LIMK2) bound to compound 52 | | Descriptor: | 4-(5-cyclopropyl-7~{H}-pyrrolo[2,3-d]pyrimidin-4-yl)-~{N}-[3-(3-methoxyphenyl)phenyl]-3,6-dihydro-2~{H}-pyridine-1-carboxamide, LIM domain kinase 2 | | Authors: | Mathea, S, Chatterjee, D, Preuss, F, Ple, K, Knapp, S. | | Deposit date: | 2024-02-20 | | Release date: | 2024-03-06 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Tetrahydropyridine LIMK inhibitors: Structure activity studies and biological characterization.
Eur.J.Med.Chem., 271, 2024
|
|
8PJA
 
 | | FKBP51FK1 F67E/K58 (i, i+9) in complex with SAFit1 | | Descriptor: | 2-[3-[(1~{R})-1-[(2~{S})-1-[(2~{S})-2-cyclohexyl-2-(3,4,5-trimethoxyphenyl)ethanoyl]piperidin-2-yl]carbonyloxy-3-(3,4-dimethoxyphenyl)propyl]phenoxy]ethanoic acid, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Charalampidou, A, Hausch, F. | | Deposit date: | 2023-06-23 | | Release date: | 2024-03-06 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Automated Flow Peptide Synthesis Enables Engineering of Proteins with Stabilized Transient Binding Pockets.
Acs Cent.Sci., 10, 2024
|
|
5VFX
 
 | | Structure of an accessory protein of the pCW3 relaxosome in complex with the origin of transfer (oriT) DNA | | Descriptor: | TcpK, oriT | | Authors: | Traore, D.A.K, Wisniewski, J.A, Flanigan, S.F, Conroy, P.J, Panjikar, S, Mok, Y.-F, Lao, C, Griffin, M.D.W, Adams, V, Rood, J.I, Whisstock, J.C. | | Deposit date: | 2017-04-10 | | Release date: | 2018-04-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Crystal structure of TcpK in complex with oriT DNA of the antibiotic resistance plasmid pCW3.
Nat Commun, 9, 2018
|
|
8VU1
 
 | | Structure of FabS1CE3-EPR-1, an elbow-locked high affinity antibody for the erythropoeitin receptor (trigonal form) | | Descriptor: | S1CE3 VARIANT OF FAB-EPR-1 heavy chain, S1CE3 VARIANT OF FAB-EPR-1 light chain | | Authors: | Singer, A.U, Bruce, H.A, Pavlenco, A, Ploder, L, Luu, G, Blazer, L, Adams, J.J, Sidhu, S.S. | | Deposit date: | 2024-01-27 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | Antigen-binding fragments with improved crystal lattice packing and enhanced conformational flexibility at the elbow region as crystallization chaperones.
Protein Sci., 33, 2024
|
|
8A5M
 
 | | TRIM7 PRYSPRY in complex with a MNV1-NS6 peptide LEALEFQ | | Descriptor: | E3 ubiquitin-protein ligase TRIM7, MNV1-NS6 peptide LEALEFQ, PHOSPHATE ION | | Authors: | Luptak, J. | | Deposit date: | 2022-06-15 | | Release date: | 2022-07-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.918 Å) | | Cite: | TRIM7 Restricts Coxsackievirus and Norovirus Infection by Detecting the C-Terminal Glutamine Generated by 3C Protease Processing.
Viruses, 14, 2022
|
|
8R1V
 
 | | Pseudomonas aeruginosa FabF C164A in complex with N-(1,5-dimethyl-3-oxo-2-phenyl-2,3-dihydro-1H-pyrazol-4-yl)-2-(4-methoxyphenoxy)acetamide | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] synthase 2, DIMETHYL SULFOXIDE, FORMIC ACID, ... | | Authors: | Yadrykhinsky, V, Brenk, R. | | Deposit date: | 2023-11-02 | | Release date: | 2024-03-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.087 Å) | | Cite: | Design, quality and validation of the EU-OPENSCREEN fragment library poised to a high-throughput screening collection.
Rsc Med Chem, 15, 2024
|
|
7Q9Q
 
 | |
5D7N
 
 | | Crystal structure of human Sirt3 at an improved resolution | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Rumpf, T, Gerhardt, S, Einsle, O, Jung, M. | | Deposit date: | 2015-08-14 | | Release date: | 2015-12-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Seeding for sirtuins: microseed matrix seeding to obtain crystals of human Sirt3 and Sirt2 suitable for soaking.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
7O2D
 
 | |
8W0O
 
 | | GDH-105 crystal structure | | Descriptor: | CHLORIDE ION, Dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Peat, T.S, Newman, J. | | Deposit date: | 2024-02-13 | | Release date: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Enhancing the Imine Reductase Activity of a Promiscuous Glucose Dehydrogenase for Scalable Manufacturing of a Chiral Neprilysin Inhibitor Precursor
Acs Catalysis, 14, 2024
|
|
5FGS
 
 | | Crystal structure of C-terminal domain of shaft pilin spaA from Lactobacillus rhamnosus GG - P21212 space group | | Descriptor: | Cell surface protein SpaA, ZINC ION | | Authors: | Chaurasia, P, Pratap, S, von Ossowski, I, Palva, A, Krishnan, V. | | Deposit date: | 2015-12-21 | | Release date: | 2016-07-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | New insights about pilus formation in gut-adapted Lactobacillus rhamnosus GG from the crystal structure of the SpaA backbone-pilin subunit
Sci Rep, 6, 2016
|
|
6NG5
 
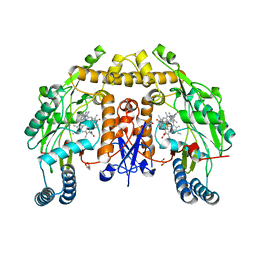 | | Structure of human neuronal nitric oxide synthase R354A/G357D mutant heme domain in complex with (R)-6-(3-fluoro-5-(2-(1-methylpyrrolidin-2-yl)ethyl)phenethyl)-4-methylpyridin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6-[2-(3-fluoro-5-{2-[(2R)-1-methylpyrrolidin-2-yl]ethyl}phenyl)ethyl]-4-methylpyridin-2-amine, GLYCEROL, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2018-12-21 | | Release date: | 2019-03-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Optimization of Blood-Brain Barrier Permeability with Potent and Selective Human Neuronal Nitric Oxide Synthase Inhibitors Having a 2-Aminopyridine Scaffold.
J. Med. Chem., 62, 2019
|
|
7NOY
 
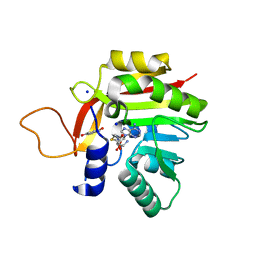 | | Crystal structure of the heterocyclic toxin methyltransferase from Mycobacterium tuberculosis in complex with substrate 1-hydroxyquinolin-4(1H)-one | | Descriptor: | 1-oxidanylquinolin-4-one, S-ADENOSYL-L-HOMOCYSTEINE, SODIUM ION, ... | | Authors: | Denkhaus, L, Sartor, P, Einsle, O, Gerhardt, S, Fetzner, S. | | Deposit date: | 2021-02-26 | | Release date: | 2021-09-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of O-methylation of (2-heptyl-)1-hydroxyquinolin-4(1H)-one and related compounds by the heterocyclic toxin methyltransferase Rv0560c of Mycobacterium tuberculosis.
J.Struct.Biol., 213, 2021
|
|
7O50
 
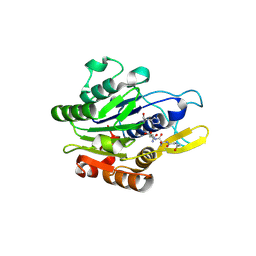 | | Crystal structure of human legumain in complex with Gly-Ser-Asn peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLY-SER-ASN, ... | | Authors: | Dall, E, Brandstetter, H. | | Deposit date: | 2021-04-07 | | Release date: | 2021-09-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Peptide Ligase Activity of Human Legumain Depends on Fold Stabilization and Balanced Substrate Affinities.
Acs Catalysis, 11, 2021
|
|
8POL
 
 | | Crystal structure of Plasmodium falciparum Sub1 protease | | Descriptor: | CALCIUM ION, PHOSPHATE ION, Subtilisin-like protease 1 | | Authors: | Martinez, M, Bouillon, A, Haouz, A, Barale, J.C, Alzari, P.M. | | Deposit date: | 2023-07-05 | | Release date: | 2024-03-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Prodomain-driven enzyme dimerization: a pH-dependent autoinhibition mechanism that controls Plasmodium Sub1 activity before merozoite egress.
Mbio, 15, 2024
|
|
6NPU
 
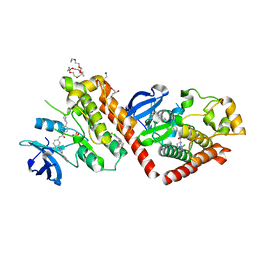 | |
7O1R
 
 | |
6NQ6
 
 | | Structure & function of a new Aspartylglucosaminuria variant | | Descriptor: | N(4)-(Beta-N-acetylglucosaminyl)-L-asparaginase | | Authors: | Pande, S, Guo, H.C. | | Deposit date: | 2019-01-19 | | Release date: | 2019-04-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The T99K variant of glycosylasparaginase shows a new structural mechanism of the genetic disease aspartylglucosaminuria.
Protein Sci., 28, 2019
|
|
7O3B
 
 | |
7ZNT
 
 | | CRYSTAL STRUCTURE OF AT7 IN COMPLEX WITH THE SECOND BROMODOMAIN OF HUMAN BRD4 AND PVHL:ELONGINC:ELONGINB | | Descriptor: | (2~{S},4~{R})-1-[(2~{R})-3-[6-[2-[(9~{S})-7-(4-chlorophenyl)-4,5,13-trimethyl-3-thia-1,8,11,12-tetrazatricyclo[8.3.0.0^{2,6}]trideca-2(6),4,7,10,12-pentaen-9-yl]ethanoylamino]hexylsulfanyl]-2-[(1-fluoranylcyclopropyl)carbonylamino]-3-methyl-butanoyl]-~{N}-[[4-(4-methyl-1,3-thiazol-5-yl)phenyl]methyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Bromodomain-containing protein 4, Elongin-B, ... | | Authors: | Hughes, S.J, Casement, R, Ciulli, A. | | Deposit date: | 2022-04-22 | | Release date: | 2022-09-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Functional E3 ligase hotspots and resistance mechanisms to small-molecule degraders.
Nat.Chem.Biol., 19, 2023
|
|
5D98
 
 | | Influenza C Virus RNA-dependent RNA Polymerase - Space group P43212 | | Descriptor: | MAGNESIUM ION, Polymerase acidic protein, Polymerase basic protein 2, ... | | Authors: | Hengrung, N, El Omari, K, Serna Martin, I, Vreede, F.T, Cusack, S, Rambo, R.P, Vonrhein, C, Bricogne, G, Stuart, D.I, Grimes, J.M, Fodor, E. | | Deposit date: | 2015-08-18 | | Release date: | 2015-10-21 | | Last modified: | 2017-09-13 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Crystal structure of the RNA-dependent RNA polymerase from influenza C virus.
Nature, 527, 2015
|
|
