4W6V
 
 | | Crystal structure of a peptide transporter from Yersinia enterocolitica at 3 A resolution | | Descriptor: | Di-/tripeptide transporter | | Authors: | Jeckelmann, J.-M, Boggavarapu, R, Harder, D, Ucurum, Z, Fotiadis, D. | | Deposit date: | 2014-08-21 | | Release date: | 2015-07-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.01819 Å) | | Cite: | Role of electrostatic interactions for ligand recognition and specificity of peptide transporters.
Bmc Biol., 13, 2015
|
|
7UOI
 
 | | Crystallographic structure of DapE from Enterococcus faecium | | Descriptor: | GLYCEROL, ZINC ION, succinyl-diaminopimelate desuccinylase | | Authors: | Gonzalez-Segura, L, Diaz-Vilchis, A, Terrazas-Lopez, M, Diaz-Sanchez, A.G. | | Deposit date: | 2022-04-12 | | Release date: | 2023-04-12 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The three-dimensional structure of DapE from Enterococcus faecium reveals new insights into DapE/ArgE subfamily ligand specificity.
Int.J.Biol.Macromol., 270, 2024
|
|
5OE3
 
 | | Crystal structure of the N-terminal domain of PqsA in complex with anthraniloyl-AMP (crystal form 1) | | Descriptor: | 1,2-ETHANEDIOL, 5'-O-[(S)-[(2-aminobenzoyl)oxy](hydroxy)phosphoryl]adenosine, ACETATE ION, ... | | Authors: | Witzgall, F, Ewert, W, Blankenfeldt, W. | | Deposit date: | 2017-07-07 | | Release date: | 2017-09-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Structures of the N-Terminal Domain of PqsA in Complex with Anthraniloyl- and 6-Fluoroanthraniloyl-AMP: Substrate Activation in Pseudomonas Quinolone Signal (PQS) Biosynthesis.
Chembiochem, 18, 2017
|
|
5OE4
 
 | |
8IC0
 
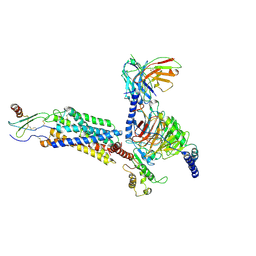 | | Cryo-EM structure of CXCL8 bound C-X-C chemokine receptor 1 in complex with Gi heterotrimer | | Descriptor: | C-X-C chemokine receptor type 1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Ishimoto, N, Park, J.H, Park, S.Y. | | Deposit date: | 2023-02-10 | | Release date: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Structural basis of CXC chemokine receptor 1 ligand binding and activation.
Nat Commun, 14, 2023
|
|
5A3S
 
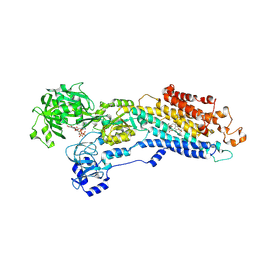 | | Crystal structure of the (SR) Calcium ATPase E2-vanadate complex bound to thapsigargin and TNP-ATP | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, OCTANOIC ACID [3S-[3ALPHA, ... | | Authors: | Clausen, J.D, Bublitz, M, Arnou, B, Olesen, C, Andersen, J.P, Moller, J.V, Nissen, P. | | Deposit date: | 2015-06-03 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal Structure of the Vanadate-Inhibited Ca(2+)-ATPase.
Structure, 24, 2016
|
|
8HIH
 
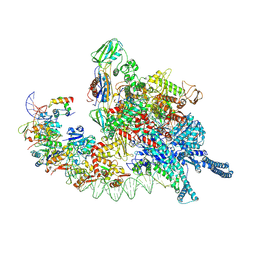 | | Cryo-EM structure of Mycobacterium tuberculosis transcription initiation complex with transcription factor GlnR | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Lin, W, Shi, J, Xu, J.C. | | Deposit date: | 2022-11-20 | | Release date: | 2023-06-07 | | Last modified: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | Structural insights into the transcription activation mechanism of the global regulator GlnR from actinobacteria.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
1C5B
 
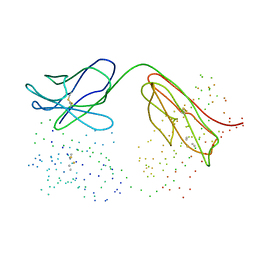 | | DECARBOXYLASE CATALYTIC ANTIBODY 21D8 UNLIGANDED FORM | | Descriptor: | CHIMERIC DECARBOXYLASE ANTIBODY 21D8 | | Authors: | Hotta, K, Wilson, I.A. | | Deposit date: | 1999-11-08 | | Release date: | 2000-10-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Catalysis of decarboxylation by a preorganized heterogeneous microenvironment: crystal structures of abzyme 21D8.
J.Mol.Biol., 302, 2000
|
|
5OE5
 
 | |
5T5U
 
 | |
3RO5
 
 | | Crystal structure of influenza A virus nucleoprotein with ligand | | Descriptor: | Nucleocapsid protein, [4-(2-chloro-4-nitrophenyl)piperazin-1-yl][3-(2-methoxyphenyl)-5-methyl-1,2-oxazol-4-yl]methanone | | Authors: | Pearce, B.C, Edavettal, S, McDonnell, P.A, Lewis, H.A, Steinbacher, S, Baldwin, E.T, Langley, D.R, Maskos, K, Mortl, M, Kiefersauer, R. | | Deposit date: | 2011-04-25 | | Release date: | 2011-09-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Inhibition of influenza virus replication via small molecules that induce the formation of higher-order nucleoprotein oligomers.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
5QTW
 
 | | FACTOR XIA IN COMPLEX WITH THE INHIBITOR methyl (2R,7S)-7-({(2E)-3-[5-chloro-2-(1H-tetrazol-1-yl)phenyl]prop-2-enoyl}amino)-14-[(methoxycarbonyl)amino]-2,3,4,5,6,7-hexahydro-1H-8,11-epimino-1,9-benzodiazacyclotridecine-2-carboxylate | | Descriptor: | 1,2-ETHANEDIOL, Coagulation factor XI, SULFATE ION, ... | | Authors: | Sheriff, S. | | Deposit date: | 2019-11-13 | | Release date: | 2020-01-29 | | Last modified: | 2021-05-12 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Orally bioavailable amine-linked macrocyclic inhibitors of factor XIa.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
5QTV
 
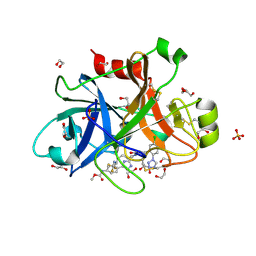 | | FACTOR XIA IN COMPLEX WITH THE INHIBITOR methyl [(2R,7S)-7-({(2E)-3-[5-chloro-2-(1H-tetrazol-1-yl)phenyl]prop-2-enoyl}amino)-2-(trifluoromethyl)-2,3,4,5,6,7-hexahydro-1H-8,11-epimino-1,9-benzodiazacyclotridecin-14-yl]carbamate | | Descriptor: | 1,2-ETHANEDIOL, Coagulation factor XI, SULFATE ION, ... | | Authors: | Sheriff, S. | | Deposit date: | 2019-11-13 | | Release date: | 2020-01-29 | | Last modified: | 2021-05-12 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Orally bioavailable amine-linked macrocyclic inhibitors of factor XIa.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
5QTX
 
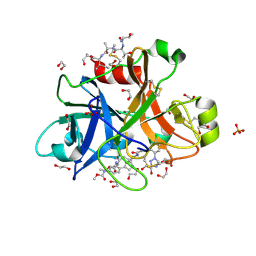 | | FACTOR XIA IN COMPLEX WITH THE INHIBITOR ethyl (2R,7S)-7-({(2E)-3-[5-chloro-2-(1H-tetrazol-1-yl)phenyl]prop-2-enoyl}amino)-14-[(methoxycarbonyl)amino]-1,2,3,4,5,6,7,9-octahydro-11,8-(azeno)-1,9-benzodiazacyclotridecine-2-carboxylate | | Descriptor: | 1,2-ETHANEDIOL, Coagulation factor XI, SULFATE ION, ... | | Authors: | Sheriff, S. | | Deposit date: | 2019-11-13 | | Release date: | 2020-01-29 | | Last modified: | 2021-05-12 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Orally bioavailable amine-linked macrocyclic inhibitors of factor XIa.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
6WJD
 
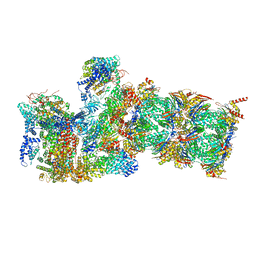 | |
5A3Q
 
 | | Crystal structure of the (SR) Calcium ATPase E2-vanadate complex bound to thapsigargin and TNP-AMPPCP | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, OCTANOIC ACID [3S-[3ALPHA, ... | | Authors: | Clausen, J.D, Bublitz, M, Arnou, B, Olesen, C, Andersen, J.P, Moller, J.V, Nissen, P. | | Deposit date: | 2015-06-02 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Crystal Structure of the Vanadate-Inhibited Ca(2+)-ATPase.
Structure, 24, 2016
|
|
6ZBT
 
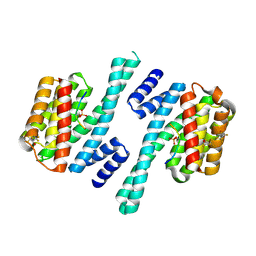 | | Structure of 14-3-3 gamma in complex with Nedd4-2 14-3-3 binding motif Ser342 | | Descriptor: | 1,1,1,3,3,3-hexafluoropropan-2-ol, 14-3-3 protein gamma, E3 ubiquitin-protein ligase NEDD4-like | | Authors: | Joshi, R, Kalabova, D, Obsil, T, Obsilova, V. | | Deposit date: | 2020-06-09 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.79948521 Å) | | Cite: | 14-3-3-protein regulates Nedd4-2 by modulating interactions between HECT and WW domains.
Commun Biol, 4, 2021
|
|
6ZC9
 
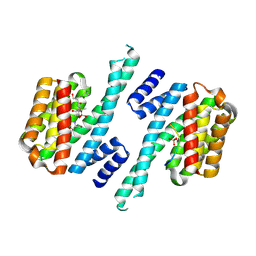 | | Structure of 14-3-3 gamma in complex with Nedd4-2 14-3-3 binding motif Ser448 | | Descriptor: | 1,1,1,3,3,3-hexafluoropropan-2-ol, 14-3-3 protein gamma, E3 ubiquitin-protein ligase NEDD4-like | | Authors: | Pohl, P, Kalabova, D, Obsil, T, Obsilova, V. | | Deposit date: | 2020-06-10 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.89895988 Å) | | Cite: | 14-3-3-protein regulates Nedd4-2 by modulating interactions between HECT and WW domains.
Commun Biol, 4, 2021
|
|
4QQI
 
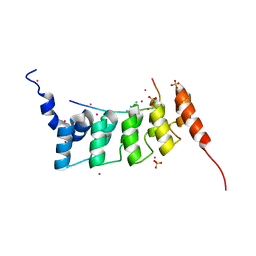 | | Crystal structure of ANKRA2-RFX7 complex | | Descriptor: | Ankyrin repeat family A protein 2, DNA-binding protein RFX7, SULFATE ION, ... | | Authors: | Xu, C, Tempel, W, Dong, A, Mackenzie, F, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-06-27 | | Release date: | 2014-09-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystal structure of ANKRA2-RFX7 complex
To Be Published
|
|
3N00
 
 | |
4ZLF
 
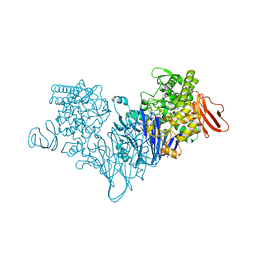 | | Cellobionic acid phosphorylase - cellobionic acid complex | | Descriptor: | 4-O-beta-D-glucopyranosyl-D-gluconic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Nam, Y.W, Arakawa, T, Fushinobu, S. | | Deposit date: | 2015-05-01 | | Release date: | 2015-06-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure and Substrate Recognition of Cellobionic Acid Phosphorylase, Which Plays a Key Role in Oxidative Cellulose Degradation by Microbes.
J.Biol.Chem., 290, 2015
|
|
3FN3
 
 | | Dimeric Structure of PD-L1 | | Descriptor: | Programmed cell death 1 ligand 1 | | Authors: | Chen, Y, Gao, F, Liu, P, Chu, F, Qi, J, Gao, G.F. | | Deposit date: | 2008-12-23 | | Release date: | 2009-12-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A dimeric structure of PD-L1: functional units or evolutionary relics?
Protein Cell, 1, 2010
|
|
1Y79
 
 | | Crystal Structure of the E.coli Dipeptidyl Carboxypeptidase Dcp in Complex with a Peptidic Inhibitor | | Descriptor: | ASPARTIC ACID, GLYCINE, LYSINE, ... | | Authors: | Comellas-Bigler, M, Lang, R, Bode, W, Maskos, K. | | Deposit date: | 2004-12-08 | | Release date: | 2005-05-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the E.coli Dipeptidyl Carboxypeptidase Dcp: Further Indication of a Ligand-dependant Hinge Movement Mechanism
J.Mol.Biol., 349, 2005
|
|
3TAW
 
 | |
6WLO
 
 | | hc16 ligase models, 11.0 Angstrom resolution | | Descriptor: | RNA (338-MER) | | Authors: | Kappel, K, Zhang, K, Su, Z, Watkins, A.M, Kladwang, W, Li, S, Pintilie, G, Topkar, V.V, Rangan, R, Zheludev, I.N, Yesselman, J.D, Chiu, W, Das, R. | | Deposit date: | 2020-04-20 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (11 Å) | | Cite: | Accelerated cryo-EM-guided determination of three-dimensional RNA-only structures.
Nat.Methods, 17, 2020
|
|
