6CGX
 
 | | Backbone cyclised conotoxin Vc1.1 mutant - D11A, E14A | | Descriptor: | Alpha-conotoxin Vc1A | | Authors: | Clark, R.J. | | Deposit date: | 2018-02-21 | | Release date: | 2018-05-23 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure-Activity Studies Reveal the Molecular Basis for GABAB-Receptor Mediated Inhibition of High Voltage-Activated Calcium Channels by alpha-Conotoxin Vc1.1.
ACS Chem. Biol., 13, 2018
|
|
6D3X
 
 | |
3C88
 
 | |
5KDH
 
 | | CRYSTAL STRUCTURE OF THE FIRST BROMODOMAIN OF HUMAN BRD4 IN COMPLEX WITH A DIHYDROPYRIDOPYRIMIDINE SCAFFOLD INHIBITOR | | Descriptor: | (5~{S})-1-ethyl-5-(4-methylphenyl)-8,9-dihydro-5~{H}-furo[3,4]pyrido[3,5-~{b}]pyrimidine-2,4,6-trione, 1,2-ETHANEDIOL, Bromodomain-containing protein 4, ... | | Authors: | Zhu, J.-Y, Schonbrunn, E. | | Deposit date: | 2016-06-08 | | Release date: | 2017-08-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | BET Bromodomain Inhibitors with One-Step Synthesis Discovered from Virtual Screen.
J. Med. Chem., 60, 2017
|
|
6KFZ
 
 | | SufS from Bacillus subtilis, soaked with L-cysteine for 90 sec at 1.96 angstrom resolution | | Descriptor: | Cysteine desulfurase SufS, DI(HYDROXYETHYL)ETHER, N-({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)-L-CYSTEINE | | Authors: | Nakamura, R, Takahashi, Y, Fujishiro, T. | | Deposit date: | 2019-07-09 | | Release date: | 2019-10-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Snapshots of PLP-substrate and PLP-product external aldimines as intermediates in two types of cysteine desulfurase enzymes.
Febs J., 287, 2020
|
|
3C8B
 
 | |
3C89
 
 | |
5KU3
 
 | | BRD4 bromodomain in complex with Cpd59 ((S)-1-(3-((2-fluoro-4-(1-methyl-1H-pyrazol-4-yl)phenyl)amino)-1-(tetrahydrofuran-3-yl)-6,7-dihydro-1H-pyrazolo[4,3-c]pyridin-5(4H)-yl)ethanone) | | Descriptor: | 1,2-ETHANEDIOL, 1-[3-[[2-fluoranyl-4-(1-methylpyrazol-4-yl)phenyl]amino]-1-[(3~{S})-oxolan-3-yl]-6,7-dihydro-4~{H}-pyrazolo[4,3-c]pyridin-5-yl]ethanone, Bromodomain-containing protein 4, ... | | Authors: | Murray, J.M, Huang, W. | | Deposit date: | 2016-07-12 | | Release date: | 2016-11-02 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Discovery of a Potent and Selective in Vivo Probe (GNE-272) for the Bromodomains of CBP/EP300.
J. Med. Chem., 59, 2016
|
|
5L41
 
 | | Thermolysin in complex with JC148 (MPD cryo protectant) | | Descriptor: | CALCIUM ION, DIMETHYL SULFOXIDE, Thermolysin, ... | | Authors: | Krimmer, S.G, Cramer, J, Heine, A, Klebe, G. | | Deposit date: | 2016-05-24 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Rational Design of Thermodynamic and Kinetic Binding Profiles by Optimizing Surface Water Networks Coating Protein-Bound Ligands.
J. Med. Chem., 59, 2016
|
|
7B7R
 
 | | MEK1 in complex with compound 4 | | Descriptor: | 2-[5-[ethyl(methyl)amino]imidazo[1,2-a]pyrimidin-7-yl]phenol, Dual specificity mitogen-activated protein kinase kinase 1,Dual specificity mitogen-activated protein kinase kinase 1, MAGNESIUM ION, ... | | Authors: | Kack, H, Oster, L. | | Deposit date: | 2020-12-11 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Fragment-Based Discovery of Novel Allosteric MEK1 Binders.
Acs Med.Chem.Lett., 12, 2021
|
|
7B3M
 
 | | MEK1 in complex with compound 6 | | Descriptor: | 1~{H}-indol-2-yl(pyridin-3-yl)methanone, Dual specificity mitogen-activated protein kinase kinase 1,Dual specificity mitogen-activated protein kinase kinase 1, MAGNESIUM ION, ... | | Authors: | Kack, H, Oster, L. | | Deposit date: | 2020-12-01 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Fragment-Based Discovery of Novel Allosteric MEK1 Binders.
Acs Med.Chem.Lett., 12, 2021
|
|
6F3B
 
 | | Crystal structure of human carbonic anhydrase I in complex with the 1-(2-hydroxy-5-sulfamoylphenyl)-3-[(4-methylphenyl)methyl]urea inhibitor | | Descriptor: | 1-[(4-methylphenyl)methyl]-3-(2-oxidanyl-5-sulfamoyl-phenyl)urea, ACETATE ION, Carbonic anhydrase 1, ... | | Authors: | Ferraroni, M, Supuran, C.T, Chiapponi, D, Bozdag, M. | | Deposit date: | 2017-11-28 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Discovery of 4-Hydroxy-3-(3-(phenylureido)benzenesulfonamides as SLC-0111 Analogues for the Treatment of Hypoxic Tumors Overexpressing Carbonic Anhydrase IX.
J. Med. Chem., 61, 2018
|
|
7NTB
 
 | |
5L8P
 
 | | Thermolysin in complex with JC114 (PEG400 cryo protectant) | | Descriptor: | CALCIUM ION, DIMETHYL SULFOXIDE, N~2~-[(R)-({[(benzyloxy)carbonyl]amino}methyl)(hydroxy)phosphoryl]-N-[(2S)-2,3,3-trimethylbutyl]-L-leucinamide, ... | | Authors: | Krimmer, S.G, Cramer, J, Heine, A, Klebe, G. | | Deposit date: | 2016-06-08 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Rational Design of Thermodynamic and Kinetic Binding Profiles by Optimizing Surface Water Networks Coating Protein-Bound Ligands.
J. Med. Chem., 59, 2016
|
|
5L3U
 
 | | Thermolysin in complex with JC149 (MPD cryo protectant) | | Descriptor: | CALCIUM ION, DIMETHYL SULFOXIDE, N~2~-[(R)-({[(benzyloxy)carbonyl]amino}methyl)(hydroxy)phosphoryl]-N-[(2R)-2,3,3-trimethylbutyl]-L-leucinamide, ... | | Authors: | Krimmer, S.G, Cramer, J, Heine, A, Klebe, G. | | Deposit date: | 2016-05-24 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Rational Design of Thermodynamic and Kinetic Binding Profiles by Optimizing Surface Water Networks Coating Protein-Bound Ligands.
J. Med. Chem., 59, 2016
|
|
8TAZ
 
 | | Cryo-EM structure of mink variant Y453F trimeric spike protein bound to one mink ACE2 receptors | | Descriptor: | Angiotensin-converting enzyme, Spike glycoprotein | | Authors: | Ahn, H.M, Calderon, B, Fan, X, Gao, Y, Horgan, N, Liang, B. | | Deposit date: | 2023-06-28 | | Release date: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Structural basis of the American mink ACE2 binding by Y453F trimeric spike glycoproteins of SARS-CoV-2.
J Med Virol, 95, 2023
|
|
8T22
 
 | | Cryo-EM structure of mink variant Y453F trimeric spike protein bound to one mink ACE2 receptors at downRBD conformation | | Descriptor: | Angiotensin-converting enzyme, Spike glycoprotein | | Authors: | Ahn, H.M, Calderon, B, Fan, X, Gao, Y, Horgan, N, Zhou, B, Liang, B. | | Deposit date: | 2023-06-05 | | Release date: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (3.83 Å) | | Cite: | Structural basis of the American mink ACE2 binding by Y453F trimeric spike glycoproteins of SARS-CoV-2.
J Med Virol, 95, 2023
|
|
8T20
 
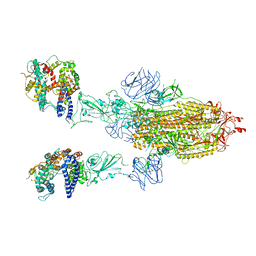 | | Cryo-EM structure of mink variant Y453F trimeric spike protein bound to two mink ACE2 receptors | | Descriptor: | Angiotensin-converting enzyme, Spike glycoprotein | | Authors: | Ahn, H.M, Calderon, B, Fan, X, Gao, Y, Horgan, N, Zhou, B, Liang, B. | | Deposit date: | 2023-06-05 | | Release date: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Structural basis of the American mink ACE2 binding by Y453F trimeric spike glycoproteins of SARS-CoV-2.
J Med Virol, 95, 2023
|
|
6V3R
 
 | | Crystal structure of murine cycloxygenase in complex with a harmaline analog, 4,9-dihydro-3H-pyrido[3,4-b]indole | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 9-[(4-chlorophenyl)methyl]-6-methoxy-1-methyl-4,9-dihydro-3H-beta-carboline, ... | | Authors: | Xu, S, Uddin, M.J, Banerjee, S, Marnett, L.J. | | Deposit date: | 2019-11-26 | | Release date: | 2020-02-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Harmaline Analogs as Substrate-Selective Cyclooxygenase-2 Inhibitors.
Acs Med.Chem.Lett., 11, 2020
|
|
5LZ8
 
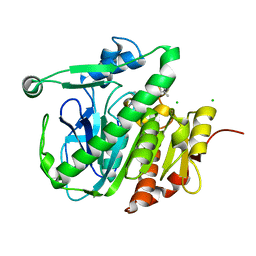 | | Fragment-based inhibitors of Lipoprotein associated Phospholipase A2 | | Descriptor: | 5-[2-[(4~{S})-4-~{tert}-butyl-2-oxidanylidene-pyrrolidin-1-yl]ethoxy]-2-fluoranyl-benzenecarbonitrile, CHLORIDE ION, Platelet-activating factor acetylhydrolase | | Authors: | Woolford, A, Day, P. | | Deposit date: | 2016-09-29 | | Release date: | 2016-12-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Fragment-Based Approach to the Development of an Orally Bioavailable Lactam Inhibitor of Lipoprotein-Associated Phospholipase A2 (Lp-PLA2).
J. Med. Chem., 59, 2016
|
|
5M3A
 
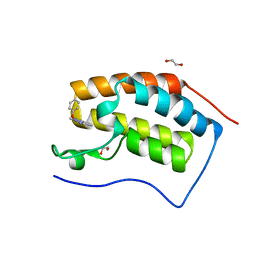 | | Crystal structure of BRD4 BROMODOMAIN 1 IN COMPLEX WITH LIGAND 2 | | Descriptor: | 1,2-ETHANEDIOL, 3-methyl-6-(1-methyl-5-phenoxy-pyrazol-4-yl)-[1,2,4]triazolo[4,3-b]pyridazine, Bromodomain-containing protein 4 | | Authors: | Kessler, D, Mayer, M, Engelhardt, H, Wolkerstorfer, B, Geist, L. | | Deposit date: | 2016-10-14 | | Release date: | 2017-09-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Direct NMR Probing of Hydration Shells of Protein Ligand Interfaces and Its Application to Drug Design.
J. Med. Chem., 60, 2017
|
|
6FAF
 
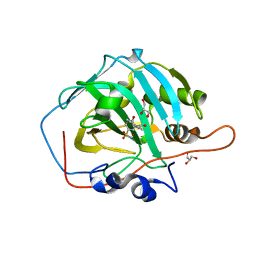 | | Crystal structure of human carbonic anhydrase I in complex with the 3-(2,5-dimethylphenyl)-1-(2-hydroxy-5-sulfamoylphenyl)urea inhibitor | | Descriptor: | 1-(2,5-dimethylphenyl)-3-(2-oxidanyl-5-sulfamoyl-phenyl)urea, Carbonic anhydrase 1, GLYCEROL, ... | | Authors: | Ferraroni, M, Supuran, C.T, Bozdag, M, Chiapponi, D. | | Deposit date: | 2017-12-15 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Discovery of 4-Hydroxy-3-(3-(phenylureido)benzenesulfonamides as SLC-0111 Analogues for the Treatment of Hypoxic Tumors Overexpressing Carbonic Anhydrase IX.
J. Med. Chem., 61, 2018
|
|
6ZU4
 
 | | Human Sirt6 13-308 in complex with ADP-ribose and the activator fluvastatin | | Descriptor: | (3R,5S,6E)-7-[3-(4-fluorophenyl)-1-(propan-2-yl)-1H-indol-2-yl]-3,5-dihydroxyhept-6-enoic acid, GLYCEROL, NAD-dependent protein deacetylase sirtuin-6, ... | | Authors: | You, W, Steegborn, C. | | Deposit date: | 2020-07-21 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Structural Basis for Activation of Human Sirtuin 6 by Fluvastatin.
Acs Med.Chem.Lett., 11, 2020
|
|
5MKZ
 
 | |
6FMD
 
 | | Targeting myeloid differentiation using potent human dihydroorotate dehydrogenase (hDHODH) inhibitors based on 2-hydroxypyrazolo[1,5-a]pyridine scaffold | | Descriptor: | 2-oxidanyl-~{N}-[2,3,5,6-tetrakis(fluoranyl)-4-phenyl-phenyl]pyrazolo[1,5-a]pyridine-3-carboxamide, ACETATE ION, CHLORIDE ION, ... | | Authors: | Goyal, P, Jarva, M, Andersson, M, Lolli, M.L, Friemann, R. | | Deposit date: | 2018-01-30 | | Release date: | 2018-07-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Targeting Myeloid Differentiation Using Potent 2-Hydroxypyrazolo[1,5- a]pyridine Scaffold-Based Human Dihydroorotate Dehydrogenase Inhibitors.
J. Med. Chem., 61, 2018
|
|
