7TE8
 
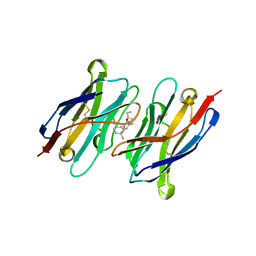 | | CA14-CBD-DB21 ternary complex | | Descriptor: | CA14, DB21, cannabidiol | | Authors: | Cao, S, Zheng, N. | | Deposit date: | 2022-01-04 | | Release date: | 2022-01-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Defining molecular glues with a dual-nanobody cannabidiol sensor.
Nat Commun, 13, 2022
|
|
1JD2
 
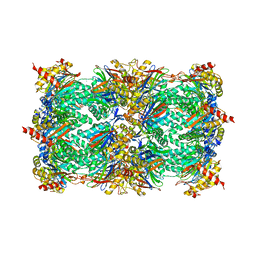 | | Crystal Structure of the yeast 20S Proteasome:TMC-95A complex: A non-covalent Proteasome Inhibitor | | Descriptor: | MAGNESIUM ION, PROTEASOME COMPONENT C1, PROTEASOME COMPONENT C11, ... | | Authors: | Groll, M, Koguchi, Y, Huber, R, Kohno, J. | | Deposit date: | 2001-06-12 | | Release date: | 2002-02-13 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of the 20 S proteasome:TMC-95A complex: a non-covalent proteasome inhibitor.
J.Mol.Biol., 311, 2001
|
|
8C1P
 
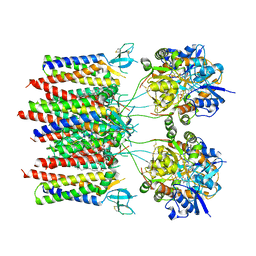 | | Active state homomeric GluA1 AMPA receptor in complex with TARP gamma 3 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, CYCLOTHIAZIDE, ... | | Authors: | Zhang, D, Ivica, J, Krieger, J.M, Ho, H, Yamashita, K, Cais, O, Greger, I. | | Deposit date: | 2022-12-21 | | Release date: | 2023-08-30 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural mobility tunes signalling of the GluA1 AMPA glutamate receptor.
Nature, 621, 2023
|
|
4S3R
 
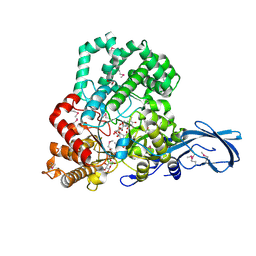 | |
6ZZX
 
 | | Structure of low-light grown Chlorella ohadii Photosystem I | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (2S)-3-{[(R)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-hydroxypropyl hexadecanoate, (3R)-beta,beta-caroten-3-ol, ... | | Authors: | Caspy, I, Nelson, N, Nechushtai, R, Neumann, E, Shkolnisky, Y. | | Deposit date: | 2020-08-05 | | Release date: | 2021-07-28 | | Last modified: | 2021-09-29 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM photosystem I structure reveals adaptation mechanisms to extreme high light in Chlorella ohadii.
Nat.Plants, 7, 2021
|
|
4CIX
 
 | | Crystal structure of Mycobacterium tuberculosis type 2 dehydroquinase in complex with(1R,4R,5R)-1,4,5-trihydroxy-3-((1S)-1-hydroxy-2-phenyl) ethylcyclohex-2-en-1-carboxylic acid | | Descriptor: | (1R,4R,5R)-1,4,5-trihydroxy-3-[(1S)-1-hydroxy-2-phenyl]ethylcyclohex-2-ene-1-carboxylic acid, 3-DEHYDROQUINATE DEHYDRATASE, SULFATE ION | | Authors: | Otero, J.M, Llamas-Saiz, A.L, Lamb, H, Hawkins, A.R, Blanco, B, Sedes, A, Peon, A, Gonzalez-Bello, C, van Raaij, M.J. | | Deposit date: | 2013-12-17 | | Release date: | 2014-04-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Exploring the water-binding pocket of the type II dehydroquinase enzyme in the structure-based design of inhibitors.
J. Med. Chem., 57, 2014
|
|
4CIY
 
 | | Crystal structure of Mycobacterium tuberculosis type 2 dehydroquinase in complex with (1R,4R,5R)-1,4,5-trihydroxy-3-((1R)-1-hydroxy-2- phenyl)ethylcyclohex-2-en-1-carboxylic acid | | Descriptor: | (1R,4R,5R)-1,4,5-trihydroxy-3-[(1R)-1-hydroxy-2-phenyl]ethylcyclohex-2-ene-1-carboxylic acid, 3-DEHYDROQUINATE DEHYDRATASE, CHLORIDE ION, ... | | Authors: | Otero, J.M, Llamas-Saiz, A.L, Lamb, H, Hawkins, A.R, Blanco, B, Sedes, A, Peon, A, Gonzalez-Bello, C, van Raaij, M.J. | | Deposit date: | 2013-12-17 | | Release date: | 2014-04-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Exploring the water-binding pocket of the type II dehydroquinase enzyme in the structure-based design of inhibitors.
J. Med. Chem., 57, 2014
|
|
7B0I
 
 | | Structure of a minimal SF3B core in complex with spliceostatin A (form II) | | Descriptor: | PHD finger-like domain-containing protein 5A, Splicing factor 3B subunit 1, Splicing factor 3B subunit 3,Splicing factor 3B subunit 3,Splicing factor 3B subunit 3, ... | | Authors: | Cretu, C, Pena, V. | | Deposit date: | 2020-11-19 | | Release date: | 2021-08-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis of intron selection by U2 snRNP in the presence of covalent inhibitors.
Nat Commun, 12, 2021
|
|
7AHR
 
 | | Enzyme of biosynthetic pathway | | Descriptor: | 3-[(1-Carboxyvinyl)oxy]benzoic acid, Chorismate dehydratase, GLYCEROL | | Authors: | Archna, A, Breithaupt, C, Stubbs, M.T. | | Deposit date: | 2020-09-25 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Mechanism of chorismate dehydratase MqnA, the first enzyme of the futalosine pathway, proceeds via substrate-assisted catalysis
J.Biol.Chem., 2022
|
|
4MNX
 
 | | Crystal structure of urokinase-type plasminogen activator (uPA) complexed with bicyclic peptide UK811 | | Descriptor: | 1,1',1''-(1,3,5-triazinane-1,3,5-triyl)tripropan-1-one, GLYCEROL, SULFATE ION, ... | | Authors: | Chen, S, Pojer, F, Heinis, C. | | Deposit date: | 2013-09-11 | | Release date: | 2014-02-05 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Peptide ligands stabilized by small molecules.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
7LX0
 
 | | Quantitative assessment of chlorophyll types in cryo-EM maps of photosystem I acclimated to far-red light | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Gisriel, C.J, Wang, J. | | Deposit date: | 2021-03-02 | | Release date: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Quantitative assessment of chlorophyll types in cryo-EM maps of photosystem I acclimated to far-red light
BBA Adv, 1, 2021
|
|
3NOY
 
 | | Crystal structure of IspG (gcpE) | | Descriptor: | 4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase, IRON/SULFUR CLUSTER | | Authors: | Groll, M, Graewert, T, Bacher, A. | | Deposit date: | 2010-06-26 | | Release date: | 2010-11-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Biosynthesis of isoprenoids: crystal structure of the [4Fe-4S] cluster protein IspG.
J.Mol.Biol., 404, 2010
|
|
1SSJ
 
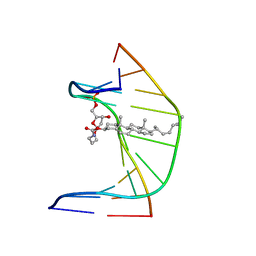 | | A DNA DUPLEX CONTAINING A CHOLESTEROL ADDUCT (BETA-ANOMER) | | Descriptor: | 5'-D(*CP*CP*AP*CP*(HOB)P*GP*GP*AP*AP*C)-3', 5'-D(GP*TP*TP*CP*CP*GP*GP*TP*GP*G)-3' | | Authors: | Gomez-Pinto, I, Cubero, E, Kalko, S.G, Monaco, V, van der Marel, G, van Boom, J.H, Orozco, M, Gonzalez, C. | | Deposit date: | 2004-03-24 | | Release date: | 2004-06-01 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Effect of bulky lesions on DNA: Solution structure of a DNA duplex containing a cholesterol adduct.
J.Biol.Chem., 279, 2004
|
|
1T27
 
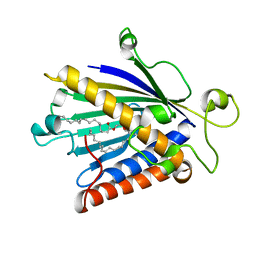 | | THE STRUCTURE OF PITP COMPLEXED TO PHOSPHATIDYLCHOLINE | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Phosphatidylinositol transfer protein alpha isoform | | Authors: | Yoder, M.D, Thomas, L.M, Tremblay, J.M, Oliver, R.L, Yarbrough, L.R, Helmkamp Jr, G.M. | | Deposit date: | 2004-04-20 | | Release date: | 2004-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | STRUCTURE OF A MULTIFUNCTIONAL PROTEIN. MAMMALIAN PHOSPHATIDYLINOSITOL TRANSFER PROTEIN COMPLEXED WITH PHOSPHATIDYLCHOLINE
J.Biol.Chem., 276, 2001
|
|
4LTC
 
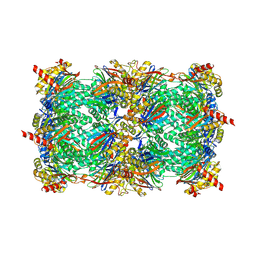 | | Crystal structure of yeast 20S proteasome in complex with enone carmaphycin analogue 6 | | Descriptor: | N-hexanoyl-L-valyl-N~1~-[(4S,5S,6R)-5-hydroxy-2,6-dimethyloctan-4-yl]-N~5~,N~5~-dimethyl-L-glutamamide, Probable proteasome subunit alpha type-7, Proteasome subunit alpha type-1, ... | | Authors: | Stein, M, Trivella, D.B.B, Groll, M. | | Deposit date: | 2013-07-23 | | Release date: | 2014-07-02 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Enzyme inhibition by hydroamination: design and mechanism of a hybrid carmaphycin-syringolin enone proteasome inhibitor.
Chem.Biol., 21, 2014
|
|
4G9P
 
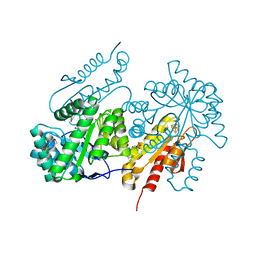 | | Structure of the GcpE-MEcPP (IspG) complex from Thermus thermophilus | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2C-METHYL-D-ERYTHRITOL 2,4-CYCLODIPHOSPHATE, 4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase, ... | | Authors: | Rekittke, I, Jomaa, H, Ermler, U. | | Deposit date: | 2012-07-24 | | Release date: | 2012-09-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure of the GcpE (IspG)-MEcPP complex from Thermus thermophilus.
Febs Lett., 586, 2012
|
|
1LBC
 
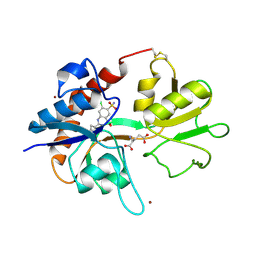 | | Crystal structure of GluR2 ligand binding core (S1S2J-N775S) in complex with cyclothiazide (CTZ) as well as glutamate at 1.8 A resolution | | Descriptor: | CYCLOTHIAZIDE, GLUTAMIC ACID, Glutamine Receptor 2, ... | | Authors: | Sun, Y, Olson, R, Horning, M, Armstrong, N, Mayer, M, Gouaux, E. | | Deposit date: | 2002-04-02 | | Release date: | 2002-05-29 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mechanism of glutamate receptor desensitization.
Nature, 417, 2002
|
|
3BW7
 
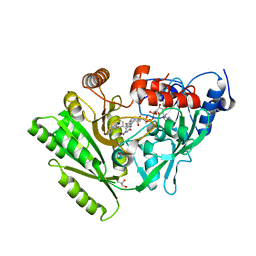 | |
3DDL
 
 | | Crystallographic Structure of Xanthorhodopsin, a Light-Driven Ion Pump with Dual Chromophore | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, RETINAL, ... | | Authors: | Stagno, J, Luecke, H, Schobert, B, Lanyi, J.K, Imasheva, E.S, Wang, J.M, Balashov, S.P. | | Deposit date: | 2008-06-05 | | Release date: | 2008-10-14 | | Last modified: | 2016-06-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic structure of xanthorhodopsin, the light-driven proton pump with a dual chromophore.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
3EL3
 
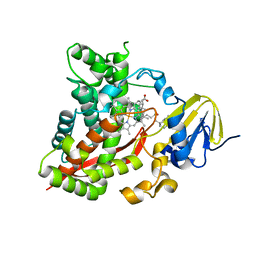 | | Distinct Monooxygenase and Farnesene Synthase Active Sites in Cytochrome P450 170A1 | | Descriptor: | (3S,3aR,6S)-3,7,7,8-tetramethyl-2,3,4,5,6,7-hexahydro-1H-3a,6-methanoazulene, PROTOPORPHYRIN IX CONTAINING FE, Putative cytochrome P450 | | Authors: | Zhao, B, Waterman, M.R. | | Deposit date: | 2008-09-19 | | Release date: | 2009-09-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structure of albaflavenone monooxygenase containing a moonlighting terpene synthase active site
J.Biol.Chem., 284, 2009
|
|
3EQL
 
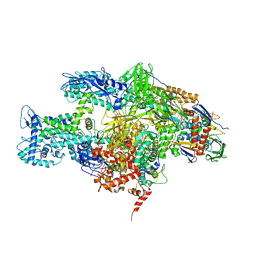 | | Crystal structure of the T. Thermophilus RNA polymerase holoenzyme in complex with antibiotic myxopyronin | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Vassylyev, D.G, Vassylyeva, M.N, Artsimovitch, I. | | Deposit date: | 2008-09-30 | | Release date: | 2008-10-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Transcription inactivation through local refolding of the RNA polymerase structure.
Nature, 457, 2009
|
|
4S3C
 
 | | IspG in complex with Epoxide Intermediate | | Descriptor: | (3S)-1,3-dihydroxy-4-{[(S)-hydroxy(phosphonooxy)phosphoryl]oxy}-2-methylbut-2-ylium, carbokation intermediate, 4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase, ... | | Authors: | Quitterer, F, Frank, A, Wang, K, Rao, G, O'Dowd, B, Li, J, Guerra, F, Abdel-Azeim, S, Bacher, A, Eppinger, J, Oldfield, E, Groll, M. | | Deposit date: | 2015-01-26 | | Release date: | 2015-05-06 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Atomic-Resolution Structures of Discrete Stages on the Reaction Coordinate of the [Fe4S4] Enzyme IspG (GcpE).
J.Mol.Biol., 427, 2015
|
|
4S3E
 
 | | IspG in complex with Inhibitor 7 (compound 1061) | | Descriptor: | 4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase, GLYCEROL, IRON/SULFUR CLUSTER, ... | | Authors: | Quitterer, F, Frank, A, Wang, K, Rao, G, O'Dowd, B, Li, J, Guerra, F, Abdel-Azeim, S, Bacher, A, Eppinger, J, Oldfield, E, Groll, M. | | Deposit date: | 2015-01-26 | | Release date: | 2015-04-29 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Atomic-Resolution Structures of Discrete Stages on the Reaction Coordinate of the [Fe4S4] Enzyme IspG (GcpE).
J.Mol.Biol., 427, 2015
|
|
4S3B
 
 | | IspG in complex with Intermediate II | | Descriptor: | (3S)-1,3-dihydroxy-4-{[(S)-hydroxy(phosphonooxy)phosphoryl]oxy}-2-methylbut-2-ylium, carbokation intermediate, 4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase, ... | | Authors: | Quitterer, F, Frank, A, Wang, K, Rao, G, O'Dowd, B, Li, J, Guerra, F, Abdel-Azeim, S, Bacher, A, Eppinger, J, Oldfield, E, Groll, M. | | Deposit date: | 2015-01-26 | | Release date: | 2015-04-29 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Atomic-Resolution Structures of Discrete Stages on the Reaction Coordinate of the [Fe4S4] Enzyme IspG (GcpE).
J.Mol.Biol., 427, 2015
|
|
4S38
 
 | | IspG in complex MEcPP | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2C-METHYL-D-ERYTHRITOL 2,4-CYCLODIPHOSPHATE, 4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase, ... | | Authors: | Quitterer, F, Frank, A, Wang, K, Guodong, R, O'Dowd, B, Li, J, Guerra, F, Abdel-Azeim, S, Bacher, A, Eppinger, J, Oldfield, E, Groll, M. | | Deposit date: | 2015-01-26 | | Release date: | 2015-04-29 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Atomic-Resolution Structures of Discrete Stages on the Reaction Coordinate of the [Fe4S4] Enzyme IspG (GcpE).
J.Mol.Biol., 427, 2015
|
|
