6KN0
 
 | |
6U1D
 
 | | Thermus thermophilus D-alanine-D-alanine ligase in complex with ATP, D-alanine-D-alanine, Mg2+ and Rb+ | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, D-ALANINE, D-alanine--D-alanine ligase, ... | | Authors: | Pederick, J.L, Bruning, J.B. | | Deposit date: | 2019-08-15 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | d-Alanine-d-alanine ligase as a model for the activation of ATP-grasp enzymes by monovalent cations.
J.Biol.Chem., 295, 2020
|
|
8PV5
 
 | | Chaetomium thermophilum pre-60S State 8 - pre-5S rotation without Foot - composite structure | | Descriptor: | 26S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Thoms, M, Cheng, J, Denk, T, Berninghausen, O, Beckmann, R. | | Deposit date: | 2023-07-17 | | Release date: | 2023-11-15 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Structural insights into coordinating 5S RNP rotation with ITS2 pre-RNA processing during ribosome formation.
Embo Rep., 24, 2023
|
|
6KL5
 
 | | Structure of The N-terminal domain of Middle East respiratory syndrome coronavirus Nucleocapsid Protein complexed with Benzyl 2-(Hydroxymethyl)-1-Indolinecarboxylate | | Descriptor: | (phenylmethyl) (2S)-2-(hydroxymethyl)-2,3-dihydroindole-1-carboxylate, Nucleoprotein | | Authors: | Hou, M.H, Lin, S.M, Hsu, J.N, Wang, Y.S. | | Deposit date: | 2019-07-29 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Structure-Based Stabilization of Non-native Protein-Protein Interactions of Coronavirus Nucleocapsid Proteins in Antiviral Drug Design.
J.Med.Chem., 63, 2020
|
|
6U1C
 
 | |
6U3D
 
 | |
6KN1
 
 | | P20/P12 of caspase-11 mutant C254A | | Descriptor: | Caspase-4 | | Authors: | Ding, J, Sun, Q. | | Deposit date: | 2019-08-02 | | Release date: | 2020-03-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Mechanism for GSDMD Targeting by Autoprocessed Caspases in Pyroptosis.
Cell, 180, 2020
|
|
6UMX
 
 | | Structural basis for specific inhibition of extracellular activation of pro/latent myostatin by SRK-015 | | Descriptor: | GL29H4-16 Fab Heavy Chain,GL29H4-16 Fab Heavy Chain, GL29H4-16 Fab Light Chain,GL29H4-16 Fab Light Chain, GLYCEROL, ... | | Authors: | Dagbay, K.B, Treece, E, Streich Jr, F.C, Jackson, J.W, Faucette, R.R, Nikiforov, A, Lin, S.C, Bostion, C.J, Nicholls, S.B, Capili, A.D, Carven, G.J. | | Deposit date: | 2019-10-10 | | Release date: | 2020-02-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structural basis of specific inhibition of extracellular activation of pro- or latent myostatin by the monoclonal antibody SRK-015.
J.Biol.Chem., 295, 2020
|
|
8PVK
 
 | | Chaetomium thermophilum pre-60S State 5 - pre-5S rotation - L1 inward - composite structure | | Descriptor: | 26S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Thoms, M, Cheng, J, Denk, T, Berninghausen, O, Beckmann, R. | | Deposit date: | 2023-07-17 | | Release date: | 2023-12-06 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (2.55 Å) | | Cite: | Structural insights into coordinating 5S RNP rotation with ITS2 pre-RNA processing during ribosome formation.
Embo Rep., 24, 2023
|
|
8PVL
 
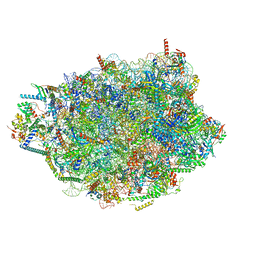 | | Chaetomium thermophilum pre-60S State 7 - pre-5S rotation lacking Utp30/ITS2 - composite structure | | Descriptor: | 26S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Thoms, M, Cheng, J, Denk, T, Berninghausen, O, Beckmann, R. | | Deposit date: | 2023-07-17 | | Release date: | 2023-12-06 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (2.19 Å) | | Cite: | Structural insights into coordinating 5S RNP rotation with ITS2 pre-RNA processing during ribosome formation.
Embo Rep., 24, 2023
|
|
6GCT
 
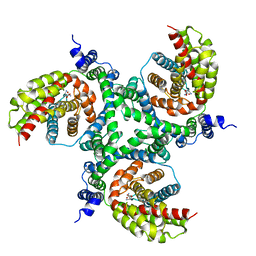 | | cryo-EM structure of the human neutral amino acid transporter ASCT2 | | Descriptor: | GLUTAMINE, Neutral amino acid transporter B(0) | | Authors: | Garaeva, A.A, Oostergetel, G.T, Gati, C, Guskov, A, Paulino, C, Slotboom, D.J. | | Deposit date: | 2018-04-19 | | Release date: | 2018-06-13 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.85 Å) | | Cite: | Cryo-EM structure of the human neutral amino acid transporter ASCT2.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6KL6
 
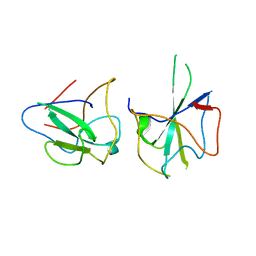 | | Crystal structure of MERS-CoV N-NTD complexed with 5-Benzyloxygramine | | Descriptor: | N,N-dimethyl-1-(5-phenylmethoxy-1H-indol-3-yl)methanamine, Nucleoprotein | | Authors: | Hou, M.H, Lin, S.M, Wang, Y.S, Hsu, J.N. | | Deposit date: | 2019-07-29 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Structure-Based Stabilization of Non-native Protein-Protein Interactions of Coronavirus Nucleocapsid Proteins in Antiviral Drug Design.
J.Med.Chem., 63, 2020
|
|
6IER
 
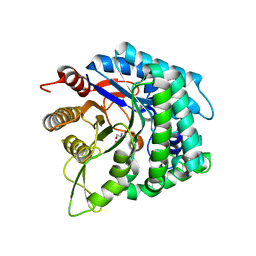 | | Apo structure of a beta-glucosidase 1317 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, beta-glucosidase 1317 | | Authors: | Xie, W, Liu, X. | | Deposit date: | 2018-09-16 | | Release date: | 2019-07-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.246 Å) | | Cite: | Improving the cellobiose-hydrolysis activity and glucose-tolerance of a thermostable beta-glucosidase through rational design.
Int.J.Biol.Macromol., 136, 2019
|
|
6IF3
 
 | | Complex structure of Rab35 and its effector ACAP2 | | Descriptor: | Arf-GAP with coiled-coil, ANK repeat and PH domain-containing protein 2, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Lin, L, Zhu, J, Zhang, R. | | Deposit date: | 2018-09-18 | | Release date: | 2019-04-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Rab35/ACAP2 and Rab35/RUSC2 Complex Structures Reveal Molecular Basis for Effector Recognition by Rab35 GTPase.
Structure, 27, 2019
|
|
6FP6
 
 | | Complex of human Cu,Zn SOD1 with the human copper chaperone for SOD1 in a compact conformation | | Descriptor: | Copper chaperone for superoxide dismutase, GLYCEROL, PROPANOIC ACID, ... | | Authors: | Sala, F.A, Wright, G.S.A, Antonyuk, S.V, Garratt, R.C, Hasnain, S.S. | | Deposit date: | 2018-02-09 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Molecular recognition and maturation of SOD1 by its evolutionarily destabilised cognate chaperone hCCS.
Plos Biol., 17, 2019
|
|
5EIL
 
 | | Computational design of a high-affinity metalloprotein homotrimer containing a metal chelating non-canonical amino acid | | Descriptor: | FE (III) ION, TRI-05 | | Authors: | Sankaran, B, Zwart, P.H, Mills, J.H, Pereira, J.H, Baker, D. | | Deposit date: | 2015-10-30 | | Release date: | 2016-11-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Computational design of a homotrimeric metalloprotein with a trisbipyridyl core.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
5EPP
 
 | | Structural Insights into the Interaction of p97 N-terminus Domain and VBM Motif in Rhomboid Protease, RHBDL4 | | Descriptor: | Rhomboid-related protein 4, Transitional endoplasmic reticulum ATPase | | Authors: | Lim, J.J, Lee, Y, Ly, T.T, Kang, J.Y, Lee, J.-G, An, J.Y, Youn, H.-S, Park, K.R, Kim, T.G, Yang, J.K, Jun, Y, Eom, S.H. | | Deposit date: | 2015-11-12 | | Release date: | 2016-09-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structural insights into the interaction of p97 N-terminus domain and VBM in rhomboid protease, RHBDL4.
Biochem.J., 473, 2016
|
|
7MU2
 
 | | Crystal Structure of WIPI2 in complex with W2IR of ATG16L1 | | Descriptor: | Autophagy-related protein 16-1, WD repeat domain phosphoinositide-interacting protein 2 | | Authors: | Strong, L.M, Flower, T.G, Buffalo, C.Z, Hurley, J.H. | | Deposit date: | 2021-05-14 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis for membrane recruitment of ATG16L1 by WIPI2 in autophagy.
Elife, 10, 2021
|
|
5EFR
 
 | |
6U1J
 
 | | Thermus thermophilus D-alanine-D-alanine ligase in complex with ADP, phosphate, D-ala-D-ala, Mg2+ and K+ | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, D-ALANINE, D-alanine--D-alanine ligase, ... | | Authors: | Pederick, J.L, Bruning, J.B, Thompson, A.P. | | Deposit date: | 2019-08-15 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | d-Alanine-d-alanine ligase as a model for the activation of ATP-grasp enzymes by monovalent cations.
J.Biol.Chem., 295, 2020
|
|
5XR3
 
 | | SAV0551 with glyoxylate | | Descriptor: | GLYOXYLIC ACID, Protein/nucleic acid deglycase HchA | | Authors: | Kim, H.J, Kwon, A.R, Lee, B.J. | | Deposit date: | 2017-06-08 | | Release date: | 2017-11-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Structural and functional studies of SAV0551 fromStaphylococcus aureusas a chaperone and glyoxalase III.
Biosci. Rep., 37, 2017
|
|
6FOL
 
 | | Domain II of the human copper chaperone in complex with human Cu,Zn superoxide dismutase | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, Copper chaperone for superoxide dismutase, GLYCEROL, ... | | Authors: | Sala, F.A, Wright, G.S.A, Antonyuk, S.V, Garratt, R.C, Hasnain, S.S. | | Deposit date: | 2018-02-07 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Molecular recognition and maturation of SOD1 by its evolutionarily destabilised cognate chaperone hCCS.
Plos Biol., 17, 2019
|
|
6FW2
 
 | | Crystal Structure of human mARC1 | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MOLYBDATE ION, Mitochondrial amidoxime-reducing component 1,Endolysin,Mitochondrial amidoxime-reducing component 1, ... | | Authors: | Kubitza, C, Scheidig, A. | | Deposit date: | 2018-03-05 | | Release date: | 2018-10-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structure of human mARC1 reveals its exceptional position among eukaryotic molybdenum enzymes.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6FWD
 
 | |
6U1E
 
 | | Thermus thermophilus D-alanine-D-alanine ligase in complex with ATP, D-alanine-D-alanine, Mg2+ and Rb+ | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, D-ALANINE, D-alanine--D-alanine ligase, ... | | Authors: | Pederick, J.L, Bruning, J.B. | | Deposit date: | 2019-08-15 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | d-Alanine-d-alanine ligase as a model for the activation of ATP-grasp enzymes by monovalent cations.
J.Biol.Chem., 295, 2020
|
|
