5K30
 
 | | Crystal structure of methionine gamma-lyase from Citrobacter freundii modified by S-Ethyl-L-cysteine sulfoxide | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, Methionine gamma-lyase, ... | | Authors: | Revtovich, S.V, Nikulin, A.D, Morozova, E.A, Demidkina, T.V. | | Deposit date: | 2016-05-19 | | Release date: | 2017-07-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Sulfoxides of sulfur-containing amino acids are suicide substrates of Citrobacter freundii methionine gamma-lyase. Structural bases of the enzyme inactivation.
Biochimie, 168, 2020
|
|
5IV7
 
 | | Cryo-electron microscopy structure of the star-shaped, hubless post-attachment T4 baseplate | | Descriptor: | Baseplate wedge protein gp10, Baseplate wedge protein gp11, Baseplate wedge protein gp25, ... | | Authors: | Taylor, N.M.I, Guerrero-Ferreira, R.C, Goldie, K.N, Stahlberg, H, Leiman, P.G. | | Deposit date: | 2016-03-19 | | Release date: | 2016-05-18 | | Last modified: | 2018-02-07 | | Method: | ELECTRON MICROSCOPY (6.77 Å) | | Cite: | Structure of the T4 baseplate and its function in triggering sheath contraction.
Nature, 533, 2016
|
|
1K70
 
 | | The Structure of Escherichia coli Cytosine Deaminase bound to 4-Hydroxy-3,4-Dihydro-1H-Pyrimidin-2-one | | Descriptor: | 4-HYDROXY-3,4-DIHYDRO-1H-PYRIMIDIN-2-ONE, Cytosine Deaminase, FE (III) ION | | Authors: | Ireton, G.C, McDermott, G, Black, M.E, Stoddard, B.L. | | Deposit date: | 2001-10-17 | | Release date: | 2002-02-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of Escherichia coli cytosine deaminase.
J.Mol.Biol., 315, 2002
|
|
1K8G
 
 | |
1KB2
 
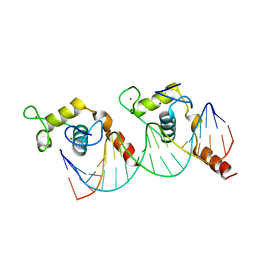 | |
1TXD
 
 | |
5JHS
 
 | |
1KHV
 
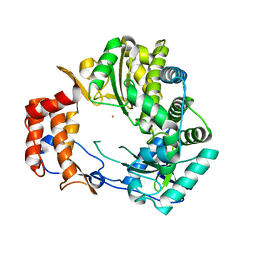 | | Crystal Structure of Rabbit Hemorrhagic Disease Virus RNA-dependent RNA polymerase complexed with Lu3+ | | Descriptor: | LUTETIUM (III) ION, RNA-DIRECTED RNA POLYMERASE | | Authors: | Ng, K.K, Cherney, M.M, Vazquez, A.L, Machin, A, Alonso, J.M, Parra, F, James, M.N. | | Deposit date: | 2001-12-01 | | Release date: | 2002-01-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of active and inactive conformations of a caliciviral RNA-dependent RNA polymerase.
J.Biol.Chem., 277, 2002
|
|
5J88
 
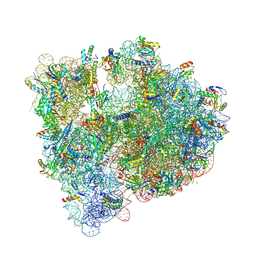 | | Structure of the E coli 70S ribosome with the U1060A mutation in 16S rRNA | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 1,4-DIAMINOBUTANE, ... | | Authors: | Cocozaki, A, Ferguson, A. | | Deposit date: | 2016-04-07 | | Release date: | 2016-07-06 | | Last modified: | 2016-12-07 | | Method: | X-RAY DIFFRACTION (3.32 Å) | | Cite: | Resistance mutations generate divergent antibiotic susceptibility profiles against translation inhibitors.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5JJ0
 
 | | Structure of G9a SET-domain with Histone H3K9M peptide and excess SAH | | Descriptor: | Histone H3K9M mutant peptide, Histone-lysine N-methyltransferase EHMT2, S-ADENOSYLMETHIONINE, ... | | Authors: | Jayaram, H, Bellon, S.F, Poy, F. | | Deposit date: | 2016-04-22 | | Release date: | 2016-07-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | S-adenosyl methionine is necessary for inhibition of the methyltransferase G9a by the lysine 9 to methionine mutation on histone H3.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5IWA
 
 | | Crystal structure of the 30S ribosomal subunit from Thermus thermophilus in complex with the GE81112 peptide antibiotic | | Descriptor: | (2S,3S)-2-{[(2S)-3-(2-amino-1H-imidazol-5-yl)-2-{[(2S,4S)-5-(carbamoyloxy)-4-hydroxy-2-({[(2S,3S)-3-hydroxypiperidin-2-yl]carbonyl}amino)pentanoyl]amino}propanoyl]amino}-3-(2-chloro-1H-imidazol-5-yl)-3-hydroxypropanoic acid, 16S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Schedlbauer, A, Kaminishi, T, Ochoa-Lizarralde, B, Chieko, N, Masahito, K, Takemoto, C, Yokoyama, S, Connell, S.R, Fucini, P. | | Deposit date: | 2016-03-22 | | Release date: | 2016-04-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Inhibition of translation initiation complex formation by GE81112 unravels a 16S rRNA structural switch involved in P-site decoding.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5IYE
 
 | | Comparison of X-ray crystal structures of a tetradecamer sequence d(CCCGGGTACCCGGG)2 at 1.7 resolution | | Descriptor: | DNA (5'-D(*CP*CP*CP*GP*GP*GP*TP*AP*CP*CP*CP*GP*GP*G)-3'), ZINC ION | | Authors: | Karthik, S, Thirugnanasambandam, A, Mandal, P.K, Gautham, N. | | Deposit date: | 2016-03-24 | | Release date: | 2017-03-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.694 Å) | | Cite: | Comparison of X-ray crystal structures of a tetradecamer sequence d(CCCGGGTACCCGGG)2 at 1.7 angstrom resolution.
Nucleosides Nucleotides Nucleic Acids, 36, 2017
|
|
5IYG
 
 | |
5IYJ
 
 | |
1U7F
 
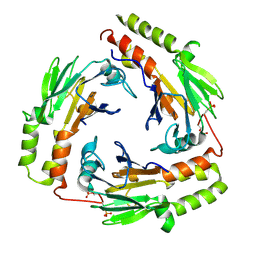 | | Crystal Structure of the phosphorylated Smad3/Smad4 heterotrimeric complex | | Descriptor: | Mothers against decapentaplegic homolog 3, Mothers against decapentaplegic homolog 4 | | Authors: | Chacko, B.M, Qin, B.Y, Tiwari, A, Shi, G, Lam, S, Hayward, L.J, de Caestecker, M, Lin, K. | | Deposit date: | 2004-08-03 | | Release date: | 2004-09-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of heteromeric smad protein assembly in tgf-Beta signaling
Mol.Cell, 15, 2004
|
|
1U7R
 
 | |
5J3W
 
 | |
1HTQ
 
 | | Multicopy crystallographic structure of a relaxed glutamine synthetase from Mycobacterium tuberculosis | | Descriptor: | ADENOSINE MONOPHOSPHATE, CITRIC ACID, MANGANESE (II) ION, ... | | Authors: | Gill, H.S, Pfluegl, G.M, Eisenberg, D, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2001-01-01 | | Release date: | 2002-07-24 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Multicopy crystallographic refinement of a relaxed glutamine synthetase from Mycobacterium tuberculosis highlights flexible loops in the enzymatic mechanism and its regulation.
Biochemistry, 41, 2002
|
|
1TMZ
 
 | | TMZIP: A CHIMERIC PEPTIDE MODEL OF THE N-TERMINUS OF ALPHA TROPOMYOSIN, NMR, 15 STRUCTURES | | Descriptor: | TMZIP | | Authors: | Greenfield, N.J, Montelione, G.T, Hitchcock-Degregori, S.E, Farid, R.S. | | Deposit date: | 1998-04-20 | | Release date: | 1998-06-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The structure of the N-terminus of striated muscle alpha-tropomyosin in a chimeric peptide: nuclear magnetic resonance structure and circular dichroism studies.
Biochemistry, 37, 1998
|
|
1TRV
 
 | |
1JZV
 
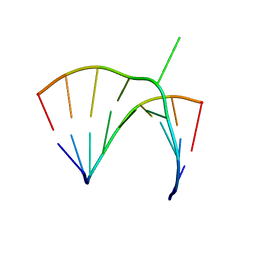 | | Crystal structure of a bulged RNA from the SL2 stem-loop of the HIV-1 psi-RNA | | Descriptor: | 5'-R(*CP*AP*GP*UP*AP*CP*GP*(5IC)P*C)-3', 5'-R(*GP*GP*CP*GP*AP*CP*(5BU)P*G)-3', MAGNESIUM ION | | Authors: | Xiong, Y, Sudarsanakumar, C, Deng, J, Pan, B, Sundaralingam, M. | | Deposit date: | 2001-09-17 | | Release date: | 2003-11-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of a Bulged RNA from the SL2 Stem-loop of the HIV-1 psi-RNA
To be Published
|
|
1JD2
 
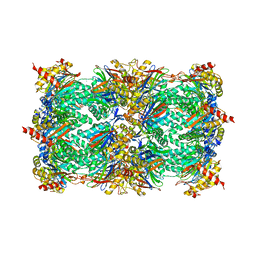 | | Crystal Structure of the yeast 20S Proteasome:TMC-95A complex: A non-covalent Proteasome Inhibitor | | Descriptor: | MAGNESIUM ION, PROTEASOME COMPONENT C1, PROTEASOME COMPONENT C11, ... | | Authors: | Groll, M, Koguchi, Y, Huber, R, Kohno, J. | | Deposit date: | 2001-06-12 | | Release date: | 2002-02-13 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of the 20 S proteasome:TMC-95A complex: a non-covalent proteasome inhibitor.
J.Mol.Biol., 311, 2001
|
|
5JQ2
 
 | |
1JJ7
 
 | |
1JJ2
 
 | | Fully Refined Crystal Structure of the Haloarcula marismortui Large Ribosomal Subunit at 2.4 Angstrom Resolution | | Descriptor: | 23S RRNA, 5S RRNA, CADMIUM ION, ... | | Authors: | Klein, D.J, Schmeing, T.M, Moore, P.B, Steitz, T.A. | | Deposit date: | 2001-07-03 | | Release date: | 2001-08-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The kink-turn: a new RNA secondary structure motif.
EMBO J., 20, 2001
|
|
