1BUF
 
 | |
2W4D
 
 | | Acylphosphatase variant G91A from Pyrococcus horikoshii | | Descriptor: | ACYLPHOSPHATASE, PHOSPHATE ION, POTASSIUM ION | | Authors: | Lam, S.Y, Wong, K.B. | | Deposit date: | 2008-11-25 | | Release date: | 2009-12-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A Rigidifying Salt-Bridge Favors the Activity of Thermophilic Enzyme at High Temperatures at the Expense of Low-Temperature Activity.
Plos Biol., 9, 2011
|
|
2W4P
 
 | | Human common-type acylphosphatase variant, A99G | | Descriptor: | ACYLPHOSPHATASE-1, GLYCEROL | | Authors: | Lam, S.Y, Sze, K.H, Wong, K.B. | | Deposit date: | 2008-11-29 | | Release date: | 2009-12-29 | | Last modified: | 2012-02-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A Rigidifying Salt-Bridge Favors the Activity of Thermophilic Enzyme at High Temperatures at the Expense of Low-Temperature Activity.
Plos Biol., 9, 2011
|
|
2W4C
 
 | | Human common-type acylphosphatase variant, A99 | | Descriptor: | ACYLPHOSPHATASE-1 | | Authors: | Lam, S.Y, Wong, K.B. | | Deposit date: | 2008-11-25 | | Release date: | 2009-12-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | A Rigidifying Salt-Bridge Favors the Activity of Thermophilic Enzyme at High Temperatures at the Expense of Low-Temperature Activity.
Plos Biol., 9, 2011
|
|
1UQB
 
 | |
1UQC
 
 | |
1UQF
 
 | |
1UQG
 
 | |
1UQE
 
 | |
1UQD
 
 | |
1UQA
 
 | |
1K2J
 
 | | NMR MINIMIZED AVERAGE STRUCTURE OF d(CGTACG)2 | | Descriptor: | 5'-D(*CP*GP*TP*AP*CP*G)-3' | | Authors: | Lam, S.L, Ip, L.N. | | Deposit date: | 2001-09-27 | | Release date: | 2002-04-10 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Low temperature solution structures and base pair stacking of double helical d(CGTACG)(2).
J.Biomol.Struct.Dyn., 19, 2002
|
|
1K2K
 
 | | NMR MINIMIZED AVERAGE STRUCTURE OF d(CGTACG)2 | | Descriptor: | 5'-D(*CP*GP*TP*AP*CP*G)-3' | | Authors: | Lam, S.L, Ip, L.N. | | Deposit date: | 2001-09-28 | | Release date: | 2002-04-10 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Low temperature solution structures and base pair stacking of double helical d(CGTACG)(2).
J.Biomol.Struct.Dyn., 19, 2002
|
|
6IY5
 
 | | NMR solution structures of 5'-ATTCTATTCT-3 | | Descriptor: | DNA (5'-D(*AP*TP*TP*CP*TP*AP*TP*TP*CP*T)-3'), SODIUM ION | | Authors: | Lam, S.L, Guo, P. | | Deposit date: | 2018-12-13 | | Release date: | 2020-06-10 | | Last modified: | 2021-09-01 | | Method: | SOLUTION NMR | | Cite: | Minidumbbell structures formed by ATTCT pentanucleotide repeats in spinocerebellar ataxia type 10.
Nucleic Acids Res., 48, 2020
|
|
6J37
 
 | | DNA minidumbbell structure of two CTTG repeats | | Descriptor: | DNA (5'-D(*CP*TP*TP*GP*CP*TP*TP*G)-3'), SODIUM ION | | Authors: | Lam, S.L, Guo, P. | | Deposit date: | 2019-01-04 | | Release date: | 2019-05-29 | | Last modified: | 2020-04-22 | | Method: | SOLUTION NMR | | Cite: | Unprecedented hydrophobic stabilizations from a reverse wobble T·T mispair in DNA minidumbbell.
J.Biomol.Struct.Dyn., 38, 2020
|
|
1DD1
 
 | |
5GWL
 
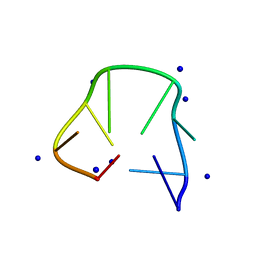 | | Structure of two CCTG repeats | | Descriptor: | DNA (5'-D(*CP*CP*TP*GP*CP*CP*TP*G)-3'), SODIUM ION | | Authors: | Guo, P, Lam, S.L. | | Deposit date: | 2016-09-12 | | Release date: | 2016-10-12 | | Method: | SOLUTION NMR | | Cite: | Minidumbbell: A New Form of Native DNA Structure
J.Am.Chem.Soc., 138, 2016
|
|
5GWQ
 
 | | Structure of two TTTA repeats | | Descriptor: | DNA (5'-D(*TP*TP*TP*AP*TP*TP*TP*A)-3'), SODIUM ION | | Authors: | Guo, P, Lam, S.L. | | Deposit date: | 2016-09-13 | | Release date: | 2016-10-12 | | Method: | SOLUTION NMR | | Cite: | Minidumbbell: A New Form of Native DNA Structure
J.Am.Chem.Soc., 138, 2016
|
|
1G88
 
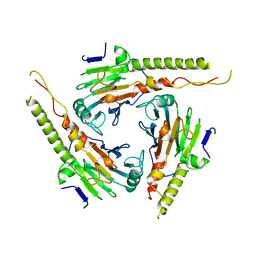 | | S4AFL3ARG515 MUTANT | | Descriptor: | SMAD4 | | Authors: | Chako, B.M, Qin, B, Lam, S.S, Correia, J.J, Lin, K. | | Deposit date: | 2000-11-16 | | Release date: | 2000-11-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The L3 loop and C-terminal phosphorylation jointly define Smad protein trimerization.
Nat.Struct.Biol., 8, 2001
|
|
2RLL
 
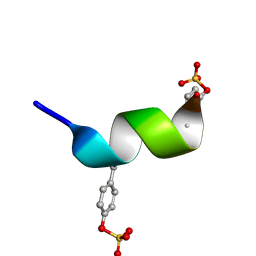 | | CCR5 Nt(7-15) | | Descriptor: | 9-mer from C-C chemokine receptor type 5 | | Authors: | Bewley, C.A, Lam, S.N. | | Deposit date: | 2007-07-21 | | Release date: | 2007-09-25 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Structures of the CCR5 N terminus and of a tyrosine-sulfated antibody with HIV-1 gp120 and CD4
Science, 317, 2007
|
|
1U7V
 
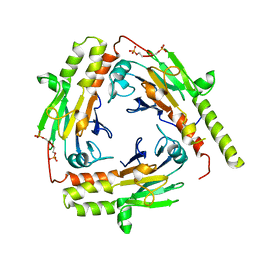 | | Crystal Structure of the phosphorylated Smad2/Smad4 heterotrimeric complex | | Descriptor: | Mothers against decapentaplegic homolog 2, Mothers against decapentaplegic homolog 4 | | Authors: | Chacko, B.M, Qin, B.Y, Tiwari, A, Shi, G, Lam, S, Hayward, L.J, de Caestecker, M, Lin, K. | | Deposit date: | 2004-08-04 | | Release date: | 2004-09-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of heteromeric smad protein assembly in tgf-Beta signaling
Mol.Cell, 15, 2004
|
|
1U7F
 
 | | Crystal Structure of the phosphorylated Smad3/Smad4 heterotrimeric complex | | Descriptor: | Mothers against decapentaplegic homolog 3, Mothers against decapentaplegic homolog 4 | | Authors: | Chacko, B.M, Qin, B.Y, Tiwari, A, Shi, G, Lam, S, Hayward, L.J, de Caestecker, M, Lin, K. | | Deposit date: | 2004-08-03 | | Release date: | 2004-09-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of heteromeric smad protein assembly in tgf-Beta signaling
Mol.Cell, 15, 2004
|
|
1MJS
 
 | |
1MK2
 
 | | SMAD3 SBD complex | | Descriptor: | ACETIC ACID, Madh-interacting protein, SMAD 3 | | Authors: | Qin, B.Y, Lam, S.S, Correia, J.J, Lin, K. | | Deposit date: | 2002-08-28 | | Release date: | 2002-10-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Smad3 allostery links TGF-beta receptor kinase activation to transcriptional control
Genes Dev., 16, 2002
|
|
7D0Z
 
 | |
