8WX0
 
 | | PNPase of M.tuberculosis with its RNA substrate | | Descriptor: | Bifunctional guanosine pentaphosphate synthetase/polyribonucleotide nucleotidyltransferase, RNA (24-mer) | | Authors: | Wang, N, Sheng, Y.N. | | Deposit date: | 2023-10-27 | | Release date: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structures of Mycobacterium tuberculosis polynucleotide phosphorylase suggest a potential mechanism for its RNA substrate degradation.
Arch.Biochem.Biophys., 754, 2024
|
|
8WWY
 
 | | Crystal structure of mouse TIFA/TIFAB heterodimer | | Descriptor: | (2R,5R)-hexane-2,5-diol, TRAF-interacting protein with FHA domain-containing protein A, TRAF-interacting protein with FHA domain-containing protein B | | Authors: | Nakamura, T. | | Deposit date: | 2023-10-27 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | TIFAB regulates the TIFA-TRAF6 signaling pathway involved in innate immunity by forming a heterodimer complex with TIFA.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8WWX
 
 | |
8WWW
 
 | | Crystal structure of the adenylation domain of SyAstC | | Descriptor: | Carrier domain-containing protein, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Li, Z, Zhu, Z, Zhu, D, Shen, Y. | | Deposit date: | 2023-10-27 | | Release date: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Functional Application of the Single-Module NRPS-like d-Alanyltransferase in Maytansinol Biosynthesis
Acs Catalysis, 14, 2024
|
|
8WWV
 
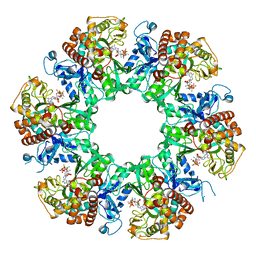 | | 1-naphthylamine GS in complex with ADP and MetSox-P | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Glutamine synthetase, L-METHIONINE-S-SULFOXIMINE PHOSPHATE, ... | | Authors: | Zhang, S.T, Zhou, N.Y. | | Deposit date: | 2023-10-26 | | Release date: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of the 1-naphthylamine biodegradation pathway reveals a glutamine synthetase-like protein that catalyzes 1-naphthylamine glutamylation
To Be Published
|
|
8WWU
 
 | | 1-naphthylamine GS in complex with AMP PNP | | Descriptor: | Glutamine synthetase, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Zhang, S.T, Zhou, N.Y. | | Deposit date: | 2023-10-26 | | Release date: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of the 1-naphthylamine biodegradation pathway reveals a glutamine synthetase-like protein that catalyzes 1-naphthylamine glutamylation
To Be Published
|
|
8WWS
 
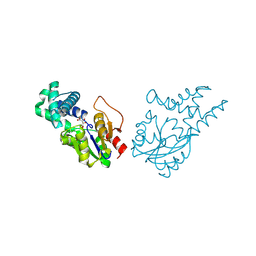 | | Crystal structure of cis-epoxysuccinate hydrolase from Klebsiella oxytoca with L(+)-tartaric acid | | Descriptor: | (S)-2-haloacid dehalogenase, 1,2-ETHANEDIOL, CARBONATE ION, ... | | Authors: | Han, Y, Kong, X.D, Li, J, Xu, J.H. | | Deposit date: | 2023-10-26 | | Release date: | 2024-06-12 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural Insights of a cis -Epoxysuccinate Hydrolase Facilitate the Development of Robust Biocatalysts for the Production of l-(+)-Tartrate.
Biochemistry, 63, 2024
|
|
8WWR
 
 | |
8WWP
 
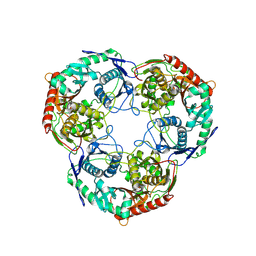 | | PNPase mutant of Mycobacterium tuberculosis | | Descriptor: | Bifunctional guanosine pentaphosphate synthetase/polyribonucleotide nucleotidyltransferase | | Authors: | Wang, N, Sheng, Y.N, Liu, Y.T. | | Deposit date: | 2023-10-26 | | Release date: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Cryo-EM structures of Mycobacterium tuberculosis polynucleotide phosphorylase suggest a potential mechanism for its RNA substrate degradation.
Arch.Biochem.Biophys., 754, 2024
|
|
8WWO
 
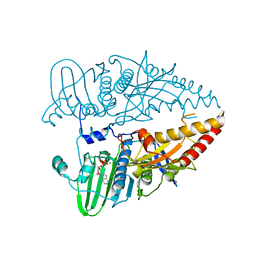 | |
8WWB
 
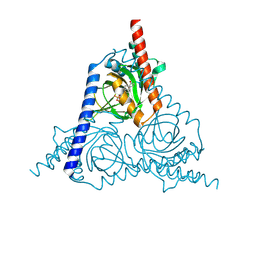 | |
8WW2
 
 | | GPR3/Gs complex | | Descriptor: | CHOLESTEROL HEMISUCCINATE, G-protein coupled receptor 3, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | He, Y, Xiong, Y. | | Deposit date: | 2023-10-24 | | Release date: | 2024-02-14 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Identification of oleic acid as an endogenous ligand of GPR3.
Cell Res., 34, 2024
|
|
8WVF
 
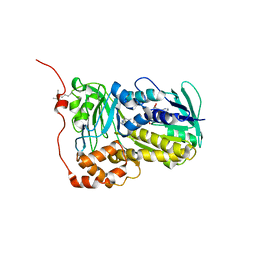 | | Crystal structure of Lsd18 mutant T189M and S195M | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Putative epoxidase LasC | | Authors: | Liu, N, Xiao, H.L, Chen, X. | | Deposit date: | 2023-10-23 | | Release date: | 2023-12-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.757 Å) | | Cite: | Simultaneous Improvement in the Thermostability and Catalytic Activity of Epoxidase Lsd18 for the Synthesis of Lasalocid A.
Int J Mol Sci, 24, 2023
|
|
8WVE
 
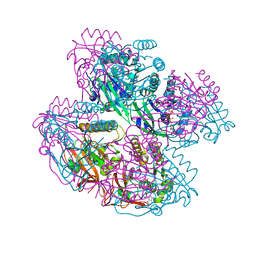 | |
8WVB
 
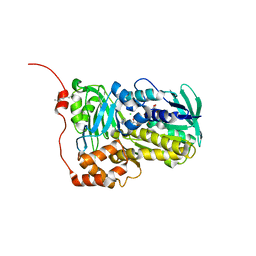 | | Crystal structure of Lsd18 mutant S195M | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, Putative epoxidase LasC | | Authors: | Liu, N, Xiao, H.L, Chen, X. | | Deposit date: | 2023-10-23 | | Release date: | 2023-12-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Simultaneous Improvement in the Thermostability and Catalytic Activity of Epoxidase Lsd18 for the Synthesis of Lasalocid A.
Int J Mol Sci, 24, 2023
|
|
8WV8
 
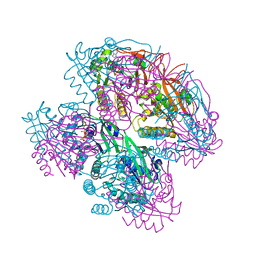 | |
8WV6
 
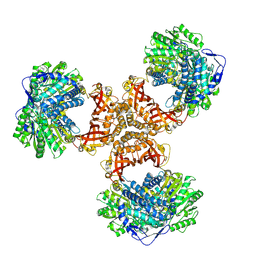 | | PaaZ, bifunctional enzyme | | Descriptor: | Bifunctional protein PaaZ | | Authors: | Yadav, S, Vinothkumar, K.R. | | Deposit date: | 2023-10-23 | | Release date: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Factors affecting macromolecule orientations in thin films formed in cryo-EM.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8WV5
 
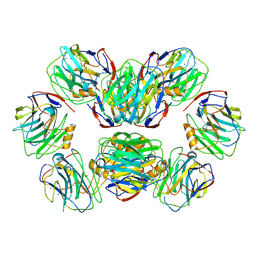 | | C-reactive protein, decamer | | Descriptor: | C-reactive protein(1-205), CALCIUM ION | | Authors: | Yadav, S, Vinothkumar, K.R. | | Deposit date: | 2023-10-23 | | Release date: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Factors affecting macromolecule orientations in thin films formed in cryo-EM.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8WV4
 
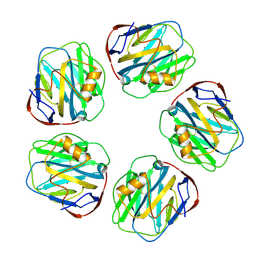 | | C-reactive protein, pentamer | | Descriptor: | C-reactive protein(1-205), CALCIUM ION | | Authors: | Yadav, S, Vinothkumar, K.R. | | Deposit date: | 2023-10-23 | | Release date: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Factors affecting macromolecule orientations in thin films formed in cryo-EM.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8WUY
 
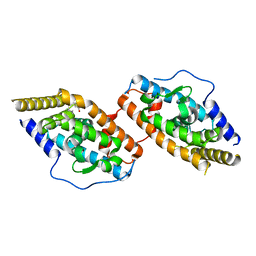 | | Crystal Structure of TR3 LBD in complex with para-positioned 3,4,5-trisubstituted benzene derivatives | | Descriptor: | Nuclear receptor subfamily 4immunitygroup A member 1, ~{N}-methyl-~{N}-octyl-3,4,5-tris(oxidanyl)benzamide | | Authors: | Hong, W.B, Chen, X.Q, Lin, T.W. | | Deposit date: | 2023-10-21 | | Release date: | 2024-01-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-based design and synthesis of anti-fibrotic compounds derived from para-positioned 3,4,5-trisubstituted benzene.
Bioorg.Chem., 144, 2024
|
|
8WUX
 
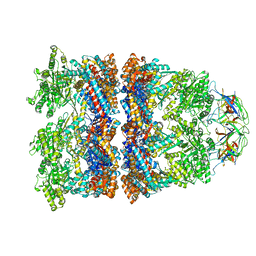 | | Cryo-EM structure of H. thermophilus GroEL-GroES bullet complex | | Descriptor: | Chaperonin GroEL, Co-chaperonin GroES, MAGNESIUM ION, ... | | Authors: | Liao, Z, Gopalasingam, C.C, Kameya, M, Gerle, C, Shigematsu, H, Ishii, M, Arakawa, T, Fushinobu, S. | | Deposit date: | 2023-10-21 | | Release date: | 2024-03-27 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural insights into thermophilic chaperonin complexes.
Structure, 32, 2024
|
|
8WUW
 
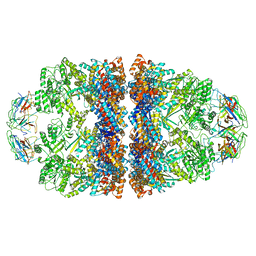 | | Cryo-EM structure of H. thermophilus GroEL-GroES2 asymmetric football complex | | Descriptor: | Chaperonin GroEL, Co-chaperonin GroES, MAGNESIUM ION, ... | | Authors: | Liao, Z, Gopalasingam, C.C, Kameya, M, Gerle, C, Shigematsu, H, Ishii, M, Arakawa, T, Fushinobu, S. | | Deposit date: | 2023-10-21 | | Release date: | 2024-03-27 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural insights into thermophilic chaperonin complexes.
Structure, 32, 2024
|
|
8WUV
 
 | | SpCas9-MMLV RT-pegRNA-target DNA complex (elongation 16-nt) | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, DNA (5'-D(*TP*GP*AP*TP*GP*GP*CP*AP*GP*AP*GP*TP*AP*CP*TP*AP*G)-3'), DNA (50-MER), ... | | Authors: | Shuto, Y, Nakagawa, R, Hoki, M, Omura, S.N, Hirano, H, Itoh, Y, Nureki, O. | | Deposit date: | 2023-10-21 | | Release date: | 2024-06-05 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for pegRNA-guided reverse transcription by a prime editor.
Nature, 631, 2024
|
|
8WUU
 
 | | SpCas9-pegRNA-target DNA complex (pre-initiation) | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, DNA (34-MER), DNA (5'-D(*TP*GP*AP*TP*GP*GP*CP*AP*GP*AP*GP*TP*AP*CP*TP*AP*G)-3'), ... | | Authors: | Shuto, Y, Nakagawa, R, Hoki, M, Omura, S.N, Hirano, H, Itoh, Y, Nureki, O. | | Deposit date: | 2023-10-21 | | Release date: | 2024-06-05 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for pegRNA-guided reverse transcription by a prime editor.
Nature, 631, 2024
|
|
8WUT
 
 | | SpCas9-MMLV RT-pegRNA-target DNA complex (initiation) | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, DNA (5'-D(*TP*GP*AP*TP*GP*GP*CP*AP*GP*AP*GP*TP*AP*CP*TP*AP*G)-3'), DNA (51-MER), ... | | Authors: | Shuto, Y, Nakagawa, R, Hoki, M, Omura, S.N, Hirano, H, Itoh, Y, Nureki, O. | | Deposit date: | 2023-10-21 | | Release date: | 2024-06-05 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for pegRNA-guided reverse transcription by a prime editor.
Nature, 631, 2024
|
|
