3IGV
 
 | | Crystal structure of HCV NS5B polymerase with a novel monocyclic dihydro-pyridinone inhibitor | | Descriptor: | N-{3-[(6S)-6-ethyl-1-(4-fluorobenzyl)-4-hydroxy-2-oxo-1,2,5,6-tetrahydropyridin-3-yl]-1,1-dioxido-2H-1,2,4-benzothiadiazin-7-yl}methanesulfonamide, RNA-DIRECTED RNA POLYMERASE | | Authors: | Zhao, Q, Showalter, R.E, Han, Q, Kissinger, C.R. | | Deposit date: | 2009-07-28 | | Release date: | 2009-12-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | 5,5'- and 6,6'-dialkyl-5,6-dihydro-1H-pyridin-2-ones as potent inhibitors of HCV NS5B polymerase.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
7LM4
 
 | | The crystal structure of the I38T mutant PA Endonuclease (2009/H1N1/CALIFORNIA) in complex with SJ000988503 | | Descriptor: | 5-hydroxy-N-[2-(4-hydroxy-3-methoxyphenyl)ethyl]-2-(2-methylphenyl)-6-oxo-1,6-dihydropyrimidine-4-carboxamide, Hexa Vinylpyrrolidone K15, MANGANESE (II) ION, ... | | Authors: | Cuypers, M.G, Slavish, P.J, Jayaraman, S, Rankovic, Z, White, S.W. | | Deposit date: | 2021-02-05 | | Release date: | 2022-02-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Chemical scaffold recycling: Structure-guided conversion of an HIV integrase inhibitor into a potent influenza virus RNA-dependent RNA polymerase inhibitor designed to minimize resistance potential.
Eur.J.Med.Chem., 247, 2023
|
|
7MTY
 
 | | The crystal structure of wild type PA endonuclease (2009/H1N1/CALIFORNIA) in complex with SJ000988569 | | Descriptor: | 2-(2,6-difluorophenyl)-5-hydroxy-N-[2-(2-methoxypyridin-4-yl)ethyl]-6-oxo-3,6-dihydropyrimidine-4-carboxamide, Hexa Vinylpyrrolidone K15, MANGANESE (II) ION, ... | | Authors: | Cuypers, M.G, Slavish, J.P, Rankovic, Z, White, S.W. | | Deposit date: | 2021-05-13 | | Release date: | 2022-05-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Chemical scaffold recycling: Structure-guided conversion of an HIV integrase inhibitor into a potent influenza virus RNA-dependent RNA polymerase inhibitor designed to minimize resistance potential.
Eur.J.Med.Chem., 247, 2023
|
|
7N47
 
 | | The crystal structure of wild type PA endonuclease (2009/H1N1/CALIFORNIA) in complex with SJ000988514 | | Descriptor: | 5-hydroxy-N-[2-(2-methoxypyridin-4-yl)ethyl]-2-(2-methylphenyl)-6-oxo-1,6-dihydropyrimidine-4-carboxamide, Hexa Vinylpyrrolidone K15, MANGANESE (II) ION, ... | | Authors: | Cuypers, M.G, Slavish, J.P, Rankovic, Z, White, S.W. | | Deposit date: | 2021-06-03 | | Release date: | 2022-06-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Chemical scaffold recycling: Structure-guided conversion of an HIV integrase inhibitor into a potent influenza virus RNA-dependent RNA polymerase inhibitor designed to minimize resistance potential.
Eur.J.Med.Chem., 247, 2023
|
|
7N55
 
 | | The crystal structure of the mutant I38T PA endonuclease (2009/H1N1/CALIFORNIA) in complex with SJ000988514 | | Descriptor: | 5-hydroxy-N-[2-(2-methoxypyridin-4-yl)ethyl]-2-(2-methylphenyl)-6-oxo-1,6-dihydropyrimidine-4-carboxamide, Hexa Vinylpyrrolidone K15, MANGANESE (II) ION, ... | | Authors: | Cuypers, M.G, Slavish, J.P, Rankovic, Z, White, S.W. | | Deposit date: | 2021-06-04 | | Release date: | 2022-06-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Chemical scaffold recycling: Structure-guided conversion of an HIV integrase inhibitor into a potent influenza virus RNA-dependent RNA polymerase inhibitor designed to minimize resistance potential.
Eur.J.Med.Chem., 247, 2023
|
|
7N8F
 
 | | The crystal structure of I38T mutant PA endonuclease (2009/H1N1/CALIFORNIA) in complex with SJ000988288 | | Descriptor: | Hexa Vinylpyrrolidone K15, MANGANESE (II) ION, Polymerase acidic protein, ... | | Authors: | Cuypers, M.G, Slavish, J.P, Rankovic, Z, White, S.W. | | Deposit date: | 2021-06-14 | | Release date: | 2022-06-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Chemical scaffold recycling: Structure-guided conversion of an HIV integrase inhibitor into a potent influenza virus RNA-dependent RNA polymerase inhibitor designed to minimize resistance potential.
Eur.J.Med.Chem., 247, 2023
|
|
2GIR
 
 | | Hepatitis C virus RNA-dependent RNA polymerase NS5B with NNI-1 inhibitor | | Descriptor: | 3-{ISOPROPYL[(TRANS-4-METHYLCYCLOHEXYL)CARBONYL]AMINO}-5-PHENYLTHIOPHENE-2-CARBOXYLIC ACID, RNA-directed RNA polymerase | | Authors: | Harris, S.F. | | Deposit date: | 2006-03-29 | | Release date: | 2007-04-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Selection and characterization of replicon variants dually resistant to thumb- and palm-binding nonnucleoside polymerase inhibitors of the hepatitis C virus.
J.Virol., 80, 2006
|
|
2GIQ
 
 | | Hepatitis C virus RNA-dependent RNA polymerase NS5B with NNI-2 inhibitor | | Descriptor: | 1-(2-CYCLOPROPYLETHYL)-3-(1,1-DIOXIDO-2H-1,2,4-BENZOTHIADIAZIN-3-YL)-6-FLUORO-4-HYDROXYQUINOLIN-2(1H)-ONE, RNA-directed RNA polymerase | | Authors: | Harris, S.F. | | Deposit date: | 2006-03-29 | | Release date: | 2007-04-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Selection and characterization of replicon variants dually resistant to thumb- and palm-binding nonnucleoside polymerase inhibitors of the hepatitis C virus.
J.Virol., 80, 2006
|
|
1ONV
 
 | | NMR Structure of a Complex Containing the TFIIF Subunit RAP74 and the RNAP II CTD Phosphatase FCP1 | | Descriptor: | Transcription initiation factor IIF, alpha subunit, serine phosphatase FCP1a | | Authors: | Nguyen, B.D, Abbott, K.L, Potempa, K, Kobor, M.S, Archambault, J, Greenblatt, J, Legault, P, Omichinski, J.G. | | Deposit date: | 2003-03-02 | | Release date: | 2003-05-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of a Complex Containing the TFIIF Subunit RAP74 and the RNA polymerase II carboxyl-terminal domain phosphatase FCP1
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
7LW6
 
 | | The crystal structure of the 2009/H1N1/California PA endonuclease I38T mutant in complex with Raltegravir | | Descriptor: | Hexa Vinylpyrrolidone K15, MANGANESE (II) ION, N-(4-fluorobenzyl)-5-hydroxy-1-methyl-2-(1-methyl-1-{[(5-methyl-1,3,4-oxadiazol-2-yl)carbonyl]amino}ethyl)-6-oxo-1,6-di hydropyrimidine-4-carboxamide, ... | | Authors: | Cuypers, M.G, Slavish, P.J, White, S.W, Rankovik, Z. | | Deposit date: | 2021-02-27 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Chemical scaffold recycling: Structure-guided conversion of an HIV integrase inhibitor into a potent influenza virus RNA-dependent RNA polymerase inhibitor designed to minimize resistance potential.
Eur.J.Med.Chem., 247, 2023
|
|
2GC8
 
 | | Structure of a Proline Sulfonamide Inhibitor Bound to HCV NS5b Polymerase | | Descriptor: | 1-[(2-AMINO-4-CHLORO-5-METHYLPHENYL)SULFONYL]-L-PROLINE, RNA-directed RNA polymerase | | Authors: | Gopalsamy, A, Chopra, R, Lim, K, Ciszewski, G, Shi, M, Curran, K.J, Sukits, S.F, Svenson, K, Bard, J, Ellingboe, J.W, Agarwal, A, Krishnamurthy, G, Howe, A.Y, Orlowski, M, Feld, B, O'connell, J, Mansour, T.S. | | Deposit date: | 2006-03-13 | | Release date: | 2006-06-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of Proline Sulfonamides as Potent and Selective Hepatitis C Virus NS5b Polymerase Inhibitors. Evidence for a New NS5b Polymerase Binding Site.
J.Med.Chem., 49, 2006
|
|
2XHV
 
 | | HCV-J4 NS5B Polymerase Point Mutant Orthorhombic Crystal Form | | Descriptor: | MAGNESIUM ION, RNA-directed RNA polymerase, SULFATE ION | | Authors: | Harrus, D, Ahmed-El-Sayed, N, Simister, P.C, Miller, S, Triconnet, M, Hagedorn, C.H, Mahias, K, Rey, F.A, Astier-Gin, T, Bressanelli, S. | | Deposit date: | 2010-06-21 | | Release date: | 2010-08-04 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Further Insights Into the Roles of GTP and the C- Terminus of the Hepatitis C Virus Polymerase in the Initiation of RNA Synthesis
J.Biol.Chem., 285, 2010
|
|
2X1B
 
 | | Structure of RNA15 RRM | | Descriptor: | MRNA 3'-END-PROCESSING PROTEIN RNA15, PHOSPHATE ION | | Authors: | Pancevac, C, Goldstone, D.C, Ramos, A, Taylor, I.A. | | Deposit date: | 2010-01-06 | | Release date: | 2010-02-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the RNA15 Rrm-RNA Complex Reveals the Molecular Basis of Gu Specificity in Transcriptional 3-End Processing Factors.
Nucleic Acids Res., 38, 2010
|
|
2R7S
 
 | | Crystal Structure of Rotavirus SA11 VP1 / RNA (UGUGCC) complex | | Descriptor: | PHOSPHATE ION, RNA (5'-R(*UP*GP*UP*GP*CP*C)-3'), RNA-dependent RNA polymerase | | Authors: | Lu, X, Harrison, S.C, Tao, Y.J, Patton, J.T, Nibert, M.L. | | Deposit date: | 2007-09-10 | | Release date: | 2008-07-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | Mechanism for coordinated RNA packaging and genome replication by rotavirus polymerase VP1.
Structure, 16, 2008
|
|
2R7X
 
 | | Crystal Structure of Rotavirus SA11 VP1/RNA (UGUGACC)/GTP complex | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, RNA (5'-R(*UP*GP*UP*GP*AP*CP*C)-3'), RNA-dependent RNA polymerase | | Authors: | Lu, X, Harrison, S.C, Tao, Y.J, Patton, J.T, Nibert, M.L. | | Deposit date: | 2007-09-10 | | Release date: | 2008-07-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mechanism for coordinated RNA packaging and genome replication by rotavirus polymerase VP1.
Structure, 16, 2008
|
|
6V6X
 
 | | The crystal structure of the 2009 H1N1 PA endonuclease mutant I38T in complex with SJ000988632 | | Descriptor: | Hexa Vinylpyrrolidone K15, MANGANESE (II) ION, N-[2-(6-amino-9H-purin-9-yl)ethyl]-5-hydroxy-6-oxo-2-[2-(trifluoromethyl)phenyl]-1,6-dihydropyrimidine-4-carboxamide, ... | | Authors: | Cuypers, M.G, Slavish, P.J, Rankovic, Z, White, S.W. | | Deposit date: | 2019-12-06 | | Release date: | 2020-12-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Chemical scaffold recycling: Structure-guided conversion of an HIV integrase inhibitor into a potent influenza virus RNA-dependent RNA polymerase inhibitor designed to minimize resistance potential.
Eur.J.Med.Chem., 247, 2023
|
|
6VG9
 
 | | The crystal structure of the 2009 H1N1/California PA endonuclease I38T mutant in complex with SJ000986248 | | Descriptor: | Hexa Vinylpyrrolidone K15, MANGANESE (II) ION, Polymerase acidic protein, ... | | Authors: | Cuypers, M.G, Slavish, P.J, Rankovic, Z, White, S.W. | | Deposit date: | 2020-01-07 | | Release date: | 2021-02-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Chemical scaffold recycling: Structure-guided conversion of an HIV integrase inhibitor into a potent influenza virus RNA-dependent RNA polymerase inhibitor designed to minimize resistance potential.
Eur.J.Med.Chem., 247, 2023
|
|
6V9E
 
 | | The crystal structure of the 2009 H1N1 PA endonuclease wild type in complex with SJ000988632 | | Descriptor: | 1,1',1'',1''',1''''-[(3R,5S,7S,9R)-decane-1,3,5,7,9-pentayl]penta(pyrrolidin-2-one), MANGANESE (II) ION, N-[2-(6-amino-9H-purin-9-yl)ethyl]-5-hydroxy-6-oxo-2-[2-(trifluoromethyl)phenyl]-1,6-dihydropyrimidine-4-carboxamide, ... | | Authors: | Cuypers, M.G, Slavish, P.J, Rankovic, Z, White, S.W. | | Deposit date: | 2019-12-13 | | Release date: | 2021-02-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Chemical scaffold recycling: Structure-guided conversion of an HIV integrase inhibitor into a potent influenza virus RNA-dependent RNA polymerase inhibitor designed to minimize resistance potential.
Eur.J.Med.Chem., 247, 2023
|
|
6VBR
 
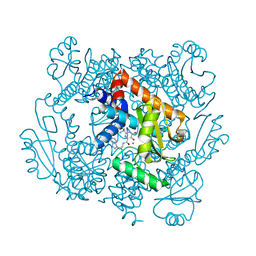 | | The crystal structure of the 2009 H1N1/California PA endonuclease wild type in complex with SJ000986248 | | Descriptor: | Hexa Vinylpyrrolidone K15, MANGANESE (II) ION, Polymerase acidic protein, ... | | Authors: | Cuypers, M.G, Slavish, P.J, Rankovic, Z, White, S.W. | | Deposit date: | 2019-12-19 | | Release date: | 2021-02-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Chemical scaffold recycling: Structure-guided conversion of an HIV integrase inhibitor into a potent influenza virus RNA-dependent RNA polymerase inhibitor designed to minimize resistance potential.
Eur.J.Med.Chem., 247, 2023
|
|
7RIL
 
 | | Crystal structure of hairpin polyamide Py-Im 1 bound to 5' CCTGACCAGG | | Descriptor: | 3-({3-[(3-{[4-({4-[(4-{[4-({(2R)-2-amino-4-[(1-methyl-4-{[1-methyl-4-({1-methyl-4-[(1-methyl-1H-imidazole-2-carbonyl)amino]-1H-imidazole-2-carbonyl}amino)-1H-pyrrole-2-carbonyl]amino}-1H-pyrrole-2-carbonyl)amino]butanoyl}amino)-1-methyl-1H-imidazole-2-carbonyl]amino}-1-methyl-1H-pyrrole-2-carbonyl)amino]-1-methyl-1H-pyrrole-2-carbonyl}amino)-1-methyl-1H-pyrrole-2-carbonyl]amino}propyl)(methyl)amino]propyl}carbamoyl)benzoic acid, ACETATE ION, non-template DNA, ... | | Authors: | Oh, J, Dervan, P.B, Wang, D. | | Deposit date: | 2021-07-20 | | Release date: | 2022-01-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | RNA polymerase II trapped on a molecular treadmill: Structural basis of persistent transcriptional arrest by a minor groove DNA binder.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6VL3
 
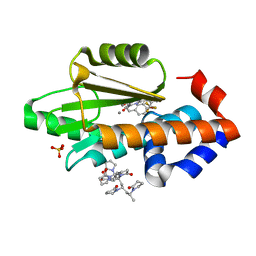 | | The crystal structure of the 2009 H1N1 PA endonuclease mutant I38T in complex with SJ000986436 | | Descriptor: | 5-hydroxy-N-[2-(4-hydroxy-3-methoxyphenyl)ethyl]-6-oxo-2-[2-(trifluoromethyl)phenyl]-1,6-dihydropyrimidine-4-carboxamide, Hexa Vinylpyrrolidone K15, MANGANESE (II) ION, ... | | Authors: | Cuypers, M.G, Slavish, P.J, Jayaraman, S, Rankovic, Z, White, S.W. | | Deposit date: | 2020-01-22 | | Release date: | 2021-02-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Chemical scaffold recycling: Structure-guided conversion of an HIV integrase inhibitor into a potent influenza virus RNA-dependent RNA polymerase inhibitor designed to minimize resistance potential.
Eur.J.Med.Chem., 247, 2023
|
|
6VJH
 
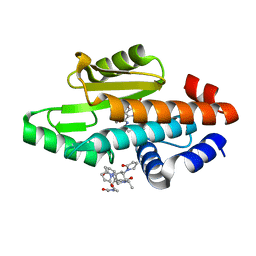 | | The crystal structure of the 2009 H1N1/California PA endonuclease wild type in complex with SJ000986192 | | Descriptor: | 2-[2-[(cyclohexylmethyl-$l^{3}-oxidanyl)carbonylamino]propan-2-yl]-~{N}-[2-(5-methoxy-4-oxidanyl-cyclohexa-1,3,5-trien-1-yl)ethyl]-5-oxidanyl-6-oxidanylidene-pyrimidine-4-carboxamide, Hexa Vinylpyrrolidone K15, MANGANESE (II) ION, ... | | Authors: | Cuypers, M.G, Slavish, P.J, Rankovic, Z, White, S.W. | | Deposit date: | 2020-01-16 | | Release date: | 2021-02-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Chemical scaffold recycling: Structure-guided conversion of an HIV integrase inhibitor into a potent influenza virus RNA-dependent RNA polymerase inhibitor designed to minimize resistance potential.
Eur.J.Med.Chem., 247, 2023
|
|
6VIV
 
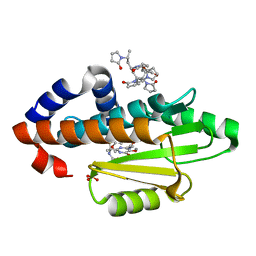 | | The crystal structure of the 2009 H1N1 PA endonuclease mutant I38T in complex with SJ000986192 | | Descriptor: | 2-[2-[(cyclohexylmethyl-$l^{3}-oxidanyl)carbonylamino]propan-2-yl]-~{N}-[2-(5-methoxy-4-oxidanyl-cyclohexa-1,3,5-trien-1-yl)ethyl]-5-oxidanyl-6-oxidanylidene-pyrimidine-4-carboxamide, Hexa Vinylpyrrolidone K15, MANGANESE (II) ION, ... | | Authors: | Cuypers, M.G, Slavish, P.J, Rankovic, Z, White, S.W. | | Deposit date: | 2020-01-14 | | Release date: | 2021-02-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Chemical scaffold recycling: Structure-guided conversion of an HIV integrase inhibitor into a potent influenza virus RNA-dependent RNA polymerase inhibitor designed to minimize resistance potential.
Eur.J.Med.Chem., 247, 2023
|
|
2XI3
 
 | | HCV-H77 NS5B Polymerase Complexed With GTP | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, RNA-directed RNA polymerase | | Authors: | Harrus, D, Ahmed-El-Sayed, N, Simister, P.C, Miller, S, Triconnet, M, Hagedorn, C.H, Mahias, K, Rey, F.A, Astier-Gin, T, Bressanelli, S. | | Deposit date: | 2010-06-25 | | Release date: | 2010-08-04 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Further Insights Into the Roles of GTP and the C- Terminus of the Hepatitis C Virus Polymerase in the Initiation of RNA Synthesis
J.Biol.Chem., 285, 2010
|
|
4AEP
 
 | |
