5U8V
 
 | | Dihydrolipoamide dehydrogenase (LpdG) from Pseudomonas aeruginosa bound to NAD+ | | Descriptor: | DIMETHYL SULFOXIDE, Dihydrolipoyl dehydrogenase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Glasser, N.R, Wang, B.X, Hoy, J.A, Newman, D.K. | | Deposit date: | 2016-12-15 | | Release date: | 2017-02-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The Pyruvate and alpha-Ketoglutarate Dehydrogenase Complexes of Pseudomonas aeruginosa Catalyze Pyocyanin and Phenazine-1-carboxylic Acid Reduction via the Subunit Dihydrolipoamide Dehydrogenase.
J. Biol. Chem., 292, 2017
|
|
6G8M
 
 | | Yeast 20S proteasome in complex with Cystargolide B Derivative 1 | | Descriptor: | (2~{S},3~{R})-4-[[(2~{S})-3-methyl-1-[[(2~{S})-3-methyl-1-oxidanylidene-1-phenylmethoxy-butan-2-yl]amino]-1-oxidanylidene-butan-2-yl]amino]-3-oxidanyl-4-oxidanylidene-2-propan-2-yl-butanoic acid, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Groll, M, Tello-Aburto, R. | | Deposit date: | 2018-04-09 | | Release date: | 2018-09-12 | | Last modified: | 2025-10-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Design, synthesis, and evaluation of cystargolide-based beta-lactones as potent proteasome inhibitors.
Eur J Med Chem, 157, 2018
|
|
7M0K
 
 | | HPK1 IN COMPLEX WITH COMPOUND 1 | | Descriptor: | 4-anilino-2-[(6-methoxy-2-methyl-1,2,3,4-tetrahydroisoquinolin-7-yl)amino]pyrimidine-5-carboxamide, Mitogen-activated protein kinase kinase kinase kinase 1 | | Authors: | Lesburg, C.A. | | Deposit date: | 2021-03-11 | | Release date: | 2021-04-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Discovery of Diaminopyrimidine Carboxamide HPK1 Inhibitors as Preclinical Immunotherapy Tool Compounds.
Acs Med.Chem.Lett., 12, 2021
|
|
5UQF
 
 | | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Campylobacter jejuni in the complex with IMP and the inhibitor P225 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Kim, Y, Maltseva, N, Makowska-Grzyska, M, Gu, M, Gollapalli, D, Hedstrom, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-08 | | Release date: | 2017-03-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from
Campylobacter jejuni in the complex with IMP and the inhibitor P225
To Be Published
|
|
7M0M
 
 | | HPK1 IN COMPLEX WITH COMPOUND 1 | | Descriptor: | 4-[2-fluoro-6-(trifluoromethyl)anilino]-2-[(6-methoxy-2-methyl-1,2,3,4-tetrahydroisoquinolin-7-yl)amino]pyrimidine-5-carboxamide, Mitogen-activated protein kinase kinase kinase kinase 1 | | Authors: | Lesburg, C.A. | | Deposit date: | 2021-03-11 | | Release date: | 2021-04-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Discovery of Diaminopyrimidine Carboxamide HPK1 Inhibitors as Preclinical Immunotherapy Tool Compounds.
Acs Med.Chem.Lett., 12, 2021
|
|
1R2Q
 
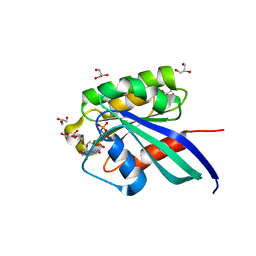 | | Crystal Structure of Human Rab5a GTPase Domain at 1.05 A resolution | | Descriptor: | GLYCEROL, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Terzyan, S, Zhu, G, Li, G, Zhang, X.C. | | Deposit date: | 2003-09-29 | | Release date: | 2003-12-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Refinement of the structure of human Rab5a GTPase domain at 1.05 A resolution.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
7M0L
 
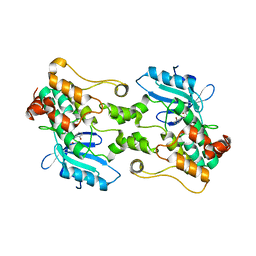 | | HPK1 IN COMPLEX WITH COMPOUND 1 | | Descriptor: | 4-(2-bromoanilino)-2-[(6-methoxy-2-methyl-1,2,3,4-tetrahydroisoquinolin-7-yl)amino]pyrimidine-5-carboxamide, Mitogen-activated protein kinase kinase kinase kinase 1 | | Authors: | Lesburg, C.A. | | Deposit date: | 2021-03-11 | | Release date: | 2021-04-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Discovery of Diaminopyrimidine Carboxamide HPK1 Inhibitors as Preclinical Immunotherapy Tool Compounds.
Acs Med.Chem.Lett., 12, 2021
|
|
2WU9
 
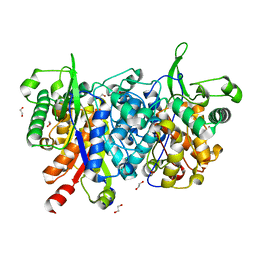 | | Crystal structure of peroxisomal KAT2 from Arabidopsis thaliana | | Descriptor: | 1,2-ETHANEDIOL, 3-KETOACYL-COA THIOLASE 2, PEROXISOMAL | | Authors: | Pye, V.E, Christensen, C.E, Dyer, J.H, Arent, S, Henriksen, A. | | Deposit date: | 2009-10-01 | | Release date: | 2010-05-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Peroxisomal Plant 3-Ketoacyl-Coa Thiolases Structure and Activity are Regulated by a Sensitive Redox Switch
J.Biol.Chem., 285, 2010
|
|
5I8F
 
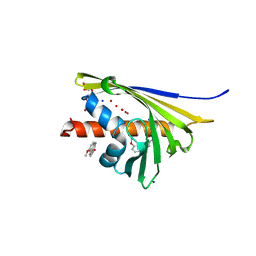 | | Crystal structure of St. John's wort Hyp-1 protein in complex with melatonin | | Descriptor: | GLYCEROL, N-[2-(5-methoxy-1H-indol-3-yl)ethyl]acetamide, Phenolic oxidative coupling protein, ... | | Authors: | Sliwiak, J, Dauter, Z, Jaskolski, M. | | Deposit date: | 2016-02-18 | | Release date: | 2016-05-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal Structure of Hyp-1, a Hypericum perforatum PR-10 Protein, in Complex with Melatonin.
Front Plant Sci, 7, 2016
|
|
7LW4
 
 | | Structure of SARS-CoV-2 nsp16/nsp10 complex in presence of S-adenosyl-L-homocysteine (SAH) | | Descriptor: | 1,2-ETHANEDIOL, 2'-O-methyltransferase, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Gupta, Y.K, Viswanathan, T, Misra, A, Qi, S. | | Deposit date: | 2021-02-27 | | Release date: | 2021-05-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A metal ion orients SARS-CoV-2 mRNA to ensure accurate 2'-O methylation of its first nucleotide.
Nat Commun, 12, 2021
|
|
5IO1
 
 | |
4COO
 
 | | Crystal structure of human cystathionine beta-synthase (delta516-525) at 2.0 angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, ACETATE ION, ... | | Authors: | McCorvie, T.J, Kopec, J, Vollamar, M, Strain-Damerell, C, Bushell, S, Bradley, A, Tallant, C, Kiyani, W, Froese, D.S, Carpenter, E.S, Burgess-Brown, N, von Delft, F, Arrowsmith, C, Edwards, A, Bountra, C, Yue, W.W. | | Deposit date: | 2014-01-29 | | Release date: | 2014-03-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Inter-Domain Communication of Human Cystathionine Beta Synthase: Structural Basis of S-Adenosyl-L-Methionine Activation.
J.Biol.Chem., 289, 2014
|
|
5ITH
 
 | |
4Q7N
 
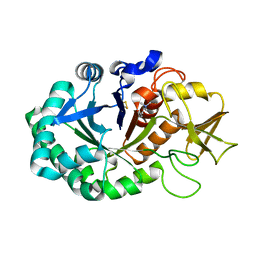 | | Crystal structure of the complex of Buffalo Signalling protein SPB-40 with 4-N-trimethylaminobutyraldehyde at 1.79 Angstrom Resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase-3-like protein 1, N,N,N-trimethyl-4-oxobutan-1-aminium | | Authors: | Chaudhary, A, Tyagi, T.K, Singh, A, Sinha, M, Bhushan, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2014-04-25 | | Release date: | 2014-05-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystal structure of the complex of Buffalo Signalling protein SPB-40 with 4-N-trimethylaminobutyraldehyde at 1.79 Angstrom Resolution
To be Published
|
|
1G2O
 
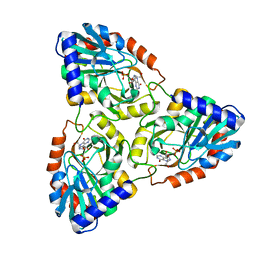 | | CRYSTAL STRUCTURE OF PURINE NUCLEOSIDE PHOSPHORYLASE FROM MYCOBACTERIUM TUBERCULOSIS IN COMPLEX WITH A TRANSITION-STATE INHIBITOR | | Descriptor: | 1,4-DIDEOXY-4-AZA-1-(S)-(9-DEAZAHYPOXANTHIN-9-YL)-D-RIBITOL, PHOSPHATE ION, PURINE NUCLEOSIDE PHOSPHORYLASE | | Authors: | Shi, W, Basso, L.A, Tyler, P.C, Furneaux, R.H, Blanchard, J.S, Almo, S.C, Schramm, V.L. | | Deposit date: | 2000-10-20 | | Release date: | 2001-08-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structures of purine nucleoside phosphorylase from Mycobacterium tuberculosis in complexes with immucillin-H and its pieces.
Biochemistry, 40, 2001
|
|
3EUY
 
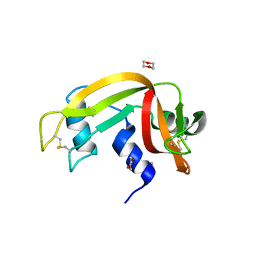 | | Crystal Structure of Ribonuclease A in 50% Dioxane | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, Ribonuclease pancreatic | | Authors: | Dechene, M, Wink, G, Smith, M, Swartz, P, Mattos, C. | | Deposit date: | 2008-10-12 | | Release date: | 2009-06-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Multiple solvent crystal structures of ribonuclease A: An assessment of the method
Proteins, 76, 2009
|
|
3A62
 
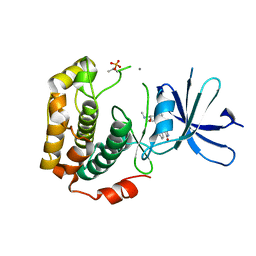 | | Crystal structure of phosphorylated p70S6K1 | | Descriptor: | MANGANESE (II) ION, Ribosomal protein S6 kinase beta-1, STAUROSPORINE | | Authors: | Sunami, T, Byrne, N, Diehl, R.E, Funabashi, K, Hall, D.L, Ikuta, M, Patel, S.B, Shipman, J.M, Smith, R.F, Takahashi, I, Zugay-Murphy, J, Iwasawa, Y, Lumb, K.J, Munshi, S.K, Sharma, S. | | Deposit date: | 2009-08-18 | | Release date: | 2009-10-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis of human p70 ribosomal S6 kinase-1 regulation by activation loop phosphorylation.
J.Biol.Chem., 285, 2010
|
|
1AVZ
 
 | |
5Z1S
 
 | | Crystal Structure Analysis of the BRD4(1) | | Descriptor: | 1,2-ETHANEDIOL, 5-bromo-2-methoxy-N-(6-methoxy-2,2-dimethyl-3-oxo-3,4-dihydro-2H-1,4-benzoxazin-7-yl)benzene-1-sulfonamide, Bromodomain-containing protein 4, ... | | Authors: | Xu, Y, Zhang, Y, Xiang, Q, Song, M, Wang, C. | | Deposit date: | 2017-12-28 | | Release date: | 2019-01-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Y08060: A Selective BET Inhibitor for Treatment of Prostate Cancer.
Acs Med.Chem.Lett., 9, 2018
|
|
4MSE
 
 | | Crystal structure of PDE10A2 with fragment ZT1597 (2-({[(2S)-2-methyl-2,3-dihydro-1,3-benzothiazol-5-yl]oxy}methyl)quinoline) | | Descriptor: | 2-{[(2-methyl-1,3-benzothiazol-5-yl)oxy]methyl}quinoline, NICKEL (II) ION, cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A | | Authors: | Sridhar, V, Badger, J, Logan, C, Chie-Leon, B, Nienaber, V. | | Deposit date: | 2013-09-18 | | Release date: | 2014-05-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Identification and optimization of PDE10A inhibitors using fragment-based screening by nanocalorimetry and X-ray crystallography.
J Biomol Screen, 19, 2014
|
|
4MW7
 
 | | Trypanosoma brucei methionyl-tRNA synthetase in complex with inhibitor 1-{3-[(5-chloro-2-ethoxy-3-iodobenzyl)amino]propyl}-3-thiophen-3-ylurea (Chem 1469) | | Descriptor: | 1-{3-[(5-chloro-2-ethoxy-3-iodobenzyl)amino]propyl}-3-thiophen-3-ylurea, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Koh, C.Y, Kim, J.E, Wetzel, A.B, de van der Schueren, W.J, Shibata, S, Liu, J, Zhang, Z, Fan, E, Verlinde, C.L.M.J, Hol, W.G.J. | | Deposit date: | 2013-09-24 | | Release date: | 2014-04-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structures of Trypanosoma brucei Methionyl-tRNA Synthetase with Urea-Based Inhibitors Provide Guidance for Drug Design against Sleeping Sickness.
Plos Negl Trop Dis, 8, 2014
|
|
6CO8
 
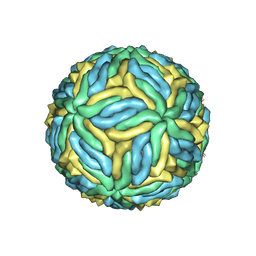 | | Structure of Zika virus at a resolution of 3.1 Angstrom | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, E protein, M protein | | Authors: | Sevvana, M, Long, F, Miller, A.J, Klose, T, Buda, G, Sun, L, Kuhn, R.J, Rossmann, M.R. | | Deposit date: | 2018-03-12 | | Release date: | 2018-07-04 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Refinement and Analysis of the Mature Zika Virus Cryo-EM Structure at 3.1 angstrom Resolution.
Structure, 26, 2018
|
|
5UPV
 
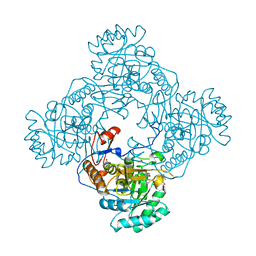 | | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Mycobacterium tuberculosis In the presence of G36 | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, INOSINIC ACID, ... | | Authors: | Kim, Y, Maltseva, N, Mulligan, R, Makowska-Grzyska, M, Gu, M, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-04 | | Release date: | 2017-02-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Mycobacterium tuberculosis In the presence of G36
To Be Published
|
|
1PN2
 
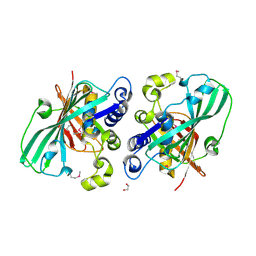 | | Crystal structure analysis of the selenomethionine labelled 2-enoyl-CoA hydratase 2 domain of Candida tropicalis multifunctional enzyme type 2 | | Descriptor: | 1,2-ETHANEDIOL, Peroxisomal hydratase-dehydrogenase-epimerase | | Authors: | Koski, M.K, Haapalainen, A.M, Hiltunen, J.K, Glumoff, T. | | Deposit date: | 2003-06-12 | | Release date: | 2004-04-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A Two-domain Structure of One Subunit Explains Unique Features of Eukaryotic Hydratase 2.
J.Biol.Chem., 279, 2004
|
|
3FVF
 
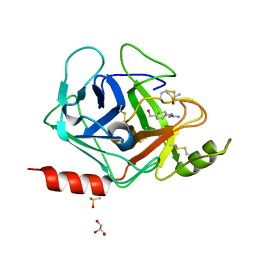 | | The Crystal Structure of Prostasin Complexed with Camostat at 1.6 Angstroms Resolution | | Descriptor: | 1-[4-(hydroxymethyl)phenyl]guanidine, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Spraggon, G, Hornsby, M, Shipway, A, Harris, J.L, Lesley, S.A. | | Deposit date: | 2009-01-15 | | Release date: | 2009-05-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Active site conformational changes of prostasin provide a new mechanism of protease regulation by divalent cations.
Protein Sci., 18, 2009
|
|
