2PHG
 
 | | Model for VP16 binding to TFIIB | | Descriptor: | Alpha trans-inducing protein, Transcription initiation factor IIB | | Authors: | Jonker, H.R.A, Wechselberger, R.W, Boelens, R, Folkers, G.E, Kaptein, R. | | Deposit date: | 2007-04-11 | | Release date: | 2007-04-24 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural Properties of the Promiscuous VP16 Activation Domain
Biochemistry, 44, 2005
|
|
5KHR
 
 | | Model of human Anaphase-promoting complex/Cyclosome complex (APC15 deletion mutant) in complex with the E2 UBE2C/UBCH10 poised for ubiquitin ligation to substrate (APC/C-CDC20-substrate-UBE2C) | | Descriptor: | Anaphase-promoting complex subunit 1, Anaphase-promoting complex subunit 10, Anaphase-promoting complex subunit 11, ... | | Authors: | VanderLinden, R, Yamaguchi, M, Dube, P, Haselbach, D, Stark, H, Schulman, B.A. | | Deposit date: | 2016-06-15 | | Release date: | 2016-08-24 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Cryo-EM of Mitotic Checkpoint Complex-Bound APC/C Reveals Reciprocal and Conformational Regulation of Ubiquitin Ligation.
Mol.Cell, 63, 2016
|
|
5LA3
 
 | | [FeFe]-hydrogenase CpI from Clostridium pasteurianum, variant E279A | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, IRON/SULFUR CLUSTER, Iron hydrogenase 1, ... | | Authors: | Duan, J, Esselborn, J, Hofmann, E, Winkler, M, Happe, T. | | Deposit date: | 2016-06-13 | | Release date: | 2017-06-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Accumulating the hydride state in the catalytic cycle of [FeFe]-hydrogenases.
Nat Commun, 8, 2017
|
|
5L9U
 
 | | Model of human Anaphase-promoting complex/Cyclosome (APC/C-CDH1) with a cross linked Ubiquitin variant-substrate-UBE2C (UBCH10) complex representing key features of multiubiquitination | | Descriptor: | Anaphase-promoting complex subunit 1, Anaphase-promoting complex subunit 10, Anaphase-promoting complex subunit 11, ... | | Authors: | Brown, N.G, VanderLinden, R, Dube, P, Haselbach, D, Peters, J.M, Stark, H, Schulman, B.A. | | Deposit date: | 2016-06-11 | | Release date: | 2016-09-14 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (6.4 Å) | | Cite: | Dual RING E3 Architectures Regulate Multiubiquitination and Ubiquitin Chain Elongation by APC/C.
Cell, 165, 2016
|
|
5L9T
 
 | | Model of human Anaphase-promoting complex/Cyclosome (APC/C-CDH1) with E2 UBE2S poised for polyubiquitination where UBE2S, APC2, and APC11 are modeled into low resolution density | | Descriptor: | Anaphase-promoting complex subunit 1, Anaphase-promoting complex subunit 10, Anaphase-promoting complex subunit 11, ... | | Authors: | Brown, N.G, VanderLinden, R, Dube, P, Haselbach, D, Peters, J.M, Stark, H, Schulman, B.A. | | Deposit date: | 2016-06-11 | | Release date: | 2016-10-26 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (6.4 Å) | | Cite: | Dual RING E3 Architectures Regulate Multiubiquitination and Ubiquitin Chain Elongation by APC/C.
Cell, 165, 2016
|
|
2R3U
 
 | | Crystal structure of the PDZ deletion mutant of DegS | | Descriptor: | Protease degS | | Authors: | Clausen, T, Kurzbauer, R. | | Deposit date: | 2007-08-30 | | Release date: | 2007-11-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Regulation of the sigmaE stress response by DegS: how the PDZ domain keeps the protease inactive in the resting state and allows integration of different OMP-derived stress signals upon folding stress.
Genes Dev., 21, 2007
|
|
5IZM
 
 | |
5IZK
 
 | |
5IZL
 
 | | The crystal structure of human eEFSec in complex with GDPCP | | Descriptor: | MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, Selenocysteine-specific elongation factor | | Authors: | Dobosz-Bartoszek, M, Simonovic, M. | | Deposit date: | 2016-03-25 | | Release date: | 2016-10-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Crystal structures of the human elongation factor eEFSec suggest a non-canonical mechanism for selenocysteine incorporation.
Nat Commun, 7, 2016
|
|
7PCS
 
 | | Structure of the heterotetrameric SDR family member BbsCD | | Descriptor: | BbsC, BbsD, GLYCEROL, ... | | Authors: | Essen, L.-O, Heider, J, von Horsten, S. | | Deposit date: | 2021-08-04 | | Release date: | 2022-06-15 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Inactive pseudoenzyme subunits in heterotetrameric BbsCD, a novel short-chain alcohol dehydrogenase involved in anaerobic toluene degradation.
Febs J., 289, 2022
|
|
6NXL
 
 | | Ubiquitin binding variants | | Descriptor: | Polyubiquitin-B | | Authors: | Miller, D.J, Watson, E.R. | | Deposit date: | 2019-02-08 | | Release date: | 2020-01-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.803 Å) | | Cite: | Protein engineering of a ubiquitin-variant inhibitor of APC/C identifies a cryptic K48 ubiquitin chain binding site.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
3FD7
 
 | | Crystal structure of Onconase C87A/C104A-ONC | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Protein P-30, ... | | Authors: | Neumann, P, Schulenburg, C, Arnold, U, Ulbrich-Hofmann, R, Stubbs, M.T. | | Deposit date: | 2008-11-25 | | Release date: | 2009-12-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.531 Å) | | Cite: | Impact of the C-terminal disulfide bond on the folding and stability of onconase.
Chembiochem, 11, 2010
|
|
4HZK
 
 | |
7UM3
 
 | | Crystal structure of a Fab in complex with a peptide derived from the LAG-3 D1 domain loop insertion | | Descriptor: | D1 domain loop peptide from Lymphocyte activation gene 3 protein, Fab heavy chain, Fab light chain | | Authors: | Zorn, J.A, Lee, P.S, Rajpal, A, Strop, P. | | Deposit date: | 2022-04-06 | | Release date: | 2022-09-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3983 Å) | | Cite: | Preclinical Characterization of Relatlimab, a Human LAG-3-Blocking Antibody, Alone or in Combination with Nivolumab.
Cancer Immunol Res, 10, 2022
|
|
3PUP
 
 | | Structure of Glycogen Synthase Kinase 3 beta (GSK3B) in complex with a ruthenium octasporine ligand (OS1) | | Descriptor: | Glycogen synthase kinase-3 beta, Ruthenium octasporine | | Authors: | Filippakopoulos, P, Kraling, K, Essen, L.O, Meggers, E, Knapp, S. | | Deposit date: | 2010-12-06 | | Release date: | 2010-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Structurally sophisticated octahedral metal complexes as highly selective protein kinase inhibitors.
J.Am.Chem.Soc., 133, 2011
|
|
6SWA
 
 | | Mus musculus brain neocortex ribosome 60S bound to Ebp1 | | Descriptor: | 28S ribosomal RNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | Authors: | Kraushar, M.L, Sprink, T. | | Deposit date: | 2019-09-20 | | Release date: | 2020-09-30 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Protein Synthesis in the Developing Neocortex at Near-Atomic Resolution Reveals Ebp1-Mediated Neuronal Proteostasis at the 60S Tunnel Exit.
Mol.Cell, 81, 2021
|
|
1DC2
 
 | | SOLUTION NMR STRUCTURE OF TUMOR SUPPRESSOR P16INK4A, 20 STRUCTURES | | Descriptor: | CYCLIN-DEPENDENT KINASE 4 INHIBITOR A (P16INK4A) | | Authors: | Byeon, I.-J.L, Li, J, Yuan, C, Tsai, M.-D. | | Deposit date: | 1999-11-04 | | Release date: | 1999-12-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Tumor suppressor INK4: refinement of p16INK4A structure and determination of p15INK4B structure by comparative modeling and NMR data.
Protein Sci., 9, 2000
|
|
1W1F
 
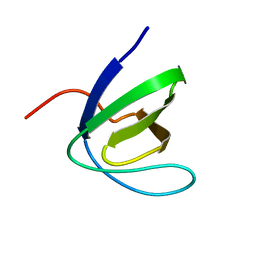 | | SH3 DOMAIN OF HUMAN LYN TYROSINE KINASE | | Descriptor: | TYROSINE-PROTEIN KINASE LYN | | Authors: | Bauer, F, Schweimer, K, Hoffmann, S, Roesch, P, Sticht, H. | | Deposit date: | 2004-06-17 | | Release date: | 2005-07-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Characterization of Lyn-SH3 Domain in Complex with a Herpesviral Protein Reveals an Extended Recognition Motif that Enhances Binding Affinity.
Protein Sci., 14, 2005
|
|
1BU9
 
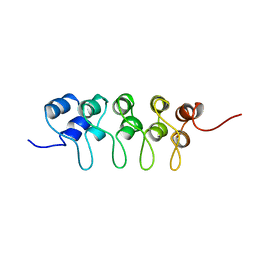 | | SOLUTION STRUCTURE OF P18-INK4C, 21 STRUCTURES | | Descriptor: | PROTEIN (CYCLIN-DEPENDENT KINASE 6 INHIBITOR) | | Authors: | Byeon, I.-J.L, Li, J, Tsai, M.-D. | | Deposit date: | 1998-09-15 | | Release date: | 1999-09-13 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Tumor suppressor INK4: determination of the solution structure of p18INK4C and demonstration of the functional significance of loops in p18INK4C and p16INK4A.
Biochemistry, 38, 1999
|
|
6U7A
 
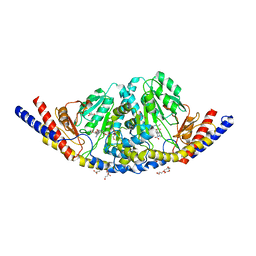 | | Rv3722c in complex with kynurenine | | Descriptor: | (2S)-4-(2-aminophenyl)-2-[(E)-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)amino]-4-oxobutanoic acid, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, 4-hydroxyquinoline-2-carboxylic acid, ... | | Authors: | Mandyoli, L, Sacchettini, J. | | Deposit date: | 2019-09-01 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Aspartate aminotransferase Rv3722c governs aspartate-dependent nitrogen metabolism in Mycobacterium tuberculosis.
Nat Commun, 11, 2020
|
|
6U78
 
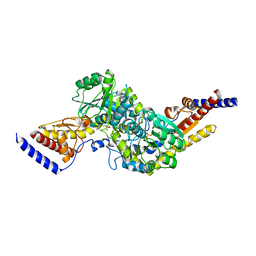 | | Rv3722c in complex with glutamic acid | | Descriptor: | Aminotransferase, GLUTAMIC ACID, GLYCEROL, ... | | Authors: | Mandyoli, L, Sacchettini, J.C. | | Deposit date: | 2019-08-31 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Aspartate aminotransferase Rv3722c governs aspartate-dependent nitrogen metabolism in Mycobacterium tuberculosis.
Nat Commun, 11, 2020
|
|
7AA4
 
 | | Structure of ClpC1-NTD bound to a CymA analogue | | Descriptor: | Negative regulator of genetic competence ClpC/mecB, polymer Cyclomarin A analogue | | Authors: | Meinhart, A, Morreale, F.E, Kaiser, M, Clausen, T. | | Deposit date: | 2020-09-03 | | Release date: | 2021-08-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | BacPROTACs mediate targeted protein degradation in bacteria.
Cell, 185, 2022
|
|
2GHF
 
 | |
7Y43
 
 | | Crystal structure of the KAT6A WH domain and its bound double stranded DNA | | Descriptor: | DNA (5'-D(*GP*GP*AP*GP*TP*GP*CP*GP*CP*AP*CP*TP*CP*C)-3'), Histone acetyltransferase KAT6A, MAGNESIUM ION | | Authors: | Wang, Z, Jia, Y. | | Deposit date: | 2022-06-13 | | Release date: | 2023-01-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The histone acetyltransferase KAT6A is recruited to unmethylated CpG islands via a DNA binding winged helix domain.
Nucleic Acids Res., 51, 2023
|
|
2GVS
 
 | | NMR solution structure of CSPsg4 | | Descriptor: | chemosensory protein CSP-sg4 | | Authors: | Tomaselli, S, Crescenzi, O, Sanfelice, D, Ab, E, Tancredi, T, Picone, D. | | Deposit date: | 2006-05-03 | | Release date: | 2006-09-12 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of a Chemosensory Protein from the Desert Locust Schistocerca gregaria(,).
Biochemistry, 45, 2006
|
|
