4HQV
 
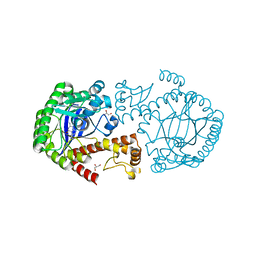 | | tRNA-guanine transglycosylase Y106F, C158V, V233G mutant in complex with queuine | | Descriptor: | 2-amino-5-({[(1S,4S,5R)-4,5-dihydroxycyclopent-2-en-1-yl]amino}methyl)-3,7-dihydro-4H-pyrrolo[2,3-d]pyrimidin-4-one, GLYCEROL, Queuine tRNA-ribosyltransferase, ... | | Authors: | Biela, I, Tidten-Luksch, N, Heine, A, Reuter, K, Klebe, G. | | Deposit date: | 2012-10-26 | | Release date: | 2012-11-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Investigation of Specificity Determinants in Bacterial tRNA-Guanine Transglycosylase Reveals Queuine, the Substrate of Its Eucaryotic Counterpart, as Inhibitor.
Plos One, 8, 2013
|
|
5JQS
 
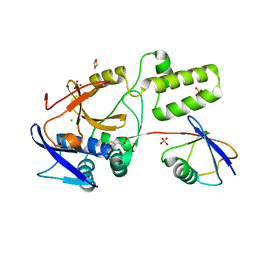 | |
7LID
 
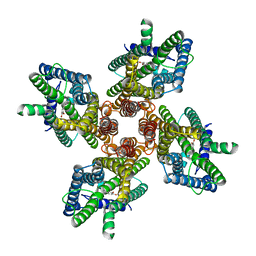 | |
7LHV
 
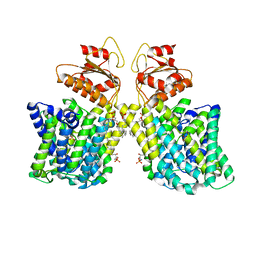 | | Structure of Arabidopsis thaliana sulfate transporter AtSULTR4;1 | | Descriptor: | (2S,3R,4E)-2-amino-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, SULFATE ION, ... | | Authors: | Wang, L, Chen, K, Zhou, M. | | Deposit date: | 2021-01-26 | | Release date: | 2021-08-11 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Structure and function of an Arabidopsis thaliana sulfate transporter.
Nat Commun, 12, 2021
|
|
7LXA
 
 | | T4 lysozyme mutant L99A | | Descriptor: | (2-methylprop-2-en-1-yl)benzene, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-MERCAPTOETHANOL, ... | | Authors: | Kamenik, A.S, Singh, I, Lak, P, Balius, T.E, Liedl, K.R, Shoichet, B.K. | | Deposit date: | 2021-03-03 | | Release date: | 2021-08-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Energy penalties enhance flexible receptor docking in a model cavity.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
4R69
 
 | | Lactate Dehydrogenase in complex with inhibitor compound 13 | | Descriptor: | (5R)-2-[(2-chlorophenyl)sulfanyl]-5-[2,6-dichloro-3-(tetrahydro-2H-pyran-4-ylamino)phenyl]-3-hydroxycyclohex-2-en-1-one, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Eigenbrot, C, Ultsch, M. | | Deposit date: | 2014-08-22 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Optimization of 5-(2,6-dichlorophenyl)-3-hydroxy-2-mercaptocyclohex-2-enones as potent inhibitors of human lactate dehydrogenase.
Bioorg.Med.Chem.Lett., 25, 2014
|
|
7LSQ
 
 | |
4R3B
 
 | |
5K2O
 
 | | Crystal structure of Arabidopsis thaliana acetohydroxyacid synthase in complex with a pyrimidinyl-benzoate herbicide, pyrithiobac | | Descriptor: | (3Z)-4-{[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]AMINO}-3-MERCAPTOPENT-3-EN-1-YL TRIHYDROGEN DIPHOSPHATE, 2-chloranyl-6-(4,6-dimethoxypyrimidin-2-yl)sulfanyl-benzoic acid, Acetolactate synthase, ... | | Authors: | Garcia, M.D, Lonhienne, T, Guddat, L.W. | | Deposit date: | 2016-05-19 | | Release date: | 2017-02-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.873 Å) | | Cite: | Comprehensive understanding of acetohydroxyacid synthase inhibition by different herbicide families.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5K3S
 
 | | Crystal structure of Arabidopsis thaliana acetohydroxyacid synthase in complex with a pyrimidinyl-benzoate herbicide, bispyribac-sodium | | Descriptor: | (3Z)-4-{[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]AMINO}-3-MERCAPTOPENT-3-EN-1-YL TRIHYDROGEN DIPHOSPHATE, 2,6-bis[(4,6-dimethoxypyrimidin-2-yl)oxy]benzoic acid, Acetolactate synthase, ... | | Authors: | Garcia, M.D, Lonhienne, T, Guddat, L.W. | | Deposit date: | 2016-05-19 | | Release date: | 2017-02-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.873 Å) | | Cite: | Comprehensive understanding of acetohydroxyacid synthase inhibition by different herbicide families.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7MTX
 
 | | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Bacillus anthracis in the complex with IMP and the inhibitor P176 | | Descriptor: | INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase, N-{2-chloro-5-[({2-[3-(prop-1-en-2-yl)phenyl]propan-2-yl}carbamoyl)amino]phenyl}-beta-D-ribopyranosylamine, ... | | Authors: | Kim, Y, Maltseva, N, Makowska-Grzyska, M, Gu, M, Gollapalli, D, Hedstrom, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-05-13 | | Release date: | 2021-06-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from
Bacillus anthracis in the complex with IMP and the inhibitor P176
To Be Published
|
|
7MC9
 
 | | X-RAY STRUCTURE OF PEDV PAPAIN-LIKE PROTEASE 2 bound to UB-PA | | Descriptor: | 3C-like proteinase, Ubiquitin, ZINC ION, ... | | Authors: | Durie, I.A, Dzimianski, J.V, Daczkowski, C.M, Pegan, S.D. | | Deposit date: | 2021-04-01 | | Release date: | 2021-07-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.099 Å) | | Cite: | Structural insights into the interaction of papain-like protease 2 from the alphacoronavirus porcine epidemic diarrhea virus and ubiquitin
Acta Cryst. D, 77, 2021
|
|
7MGY
 
 | | Sco GlgEI-V279S in complex with 4-alpha-glucoside of valienamine | | Descriptor: | (1R,4S,5S,6R)-4-amino-5,6-dihydroxy-2-(hydroxymethyl)cyclohex-2-en-1-yl alpha-D-glucopyranoside, Alpha-1,4-glucan:maltose-1-phosphate maltosyltransferase 1, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Jayasinghe, T.D, Ronning, D.R. | | Deposit date: | 2021-04-13 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Stereoselective synthesis of a 4-⍺-glucoside of valienamine and its X-ray structure in complex with Streptomyces coelicolor GlgE1-V279S.
Sci Rep, 11, 2021
|
|
7MLM
 
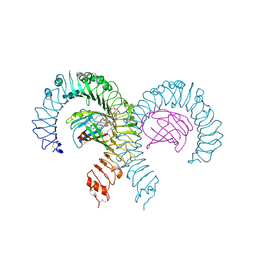 | | Crystal structure of mouse TLR4/MD-2 in complex with sulfatides | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Su, L, Beutler, B. | | Deposit date: | 2021-04-28 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.104 Å) | | Cite: | Sulfatides are endogenous ligands for the TLR4-MD-2 complex.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
5KDI
 
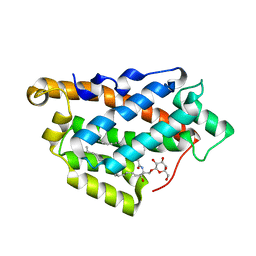 | | How FAPP2 Selects Simple Glycosphingolipids Using the GLTP-fold | | Descriptor: | (~{Z})-~{N}-[(~{E},2~{S},3~{R})-1-[(2~{R},3~{R},4~{S},5~{R},6~{R})-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-3-oxidanyl-octadec-4-en-2-yl]octadec-9-enamide, Pleckstrin homology domain-containing family A member 8 | | Authors: | Ochoa-Lizarralde, B, Popov, A.N, Samygina, V.R, Patel, D.J, Brown, R.E, Malinina, L. | | Deposit date: | 2016-06-08 | | Release date: | 2017-12-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural analyses of 4-phosphate adaptor protein 2 yield mechanistic insights into sphingolipid recognition by the glycolipid transfer protein family.
J.Biol.Chem., 293, 2018
|
|
4RN2
 
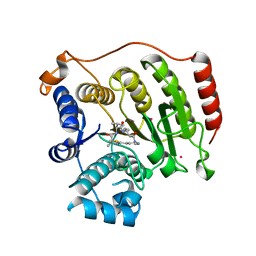 | | Crystal structure of S39D HDAC8 in complex with a largazole analogue. | | Descriptor: | (5R,8S,11S)-5-methyl-8-(propan-2-yl)-11-[(1E)-4-sulfanylbut-1-en-1-yl]-3-thia-7,10,14,17,21-pentaazatricyclo[14.3.1.1~2,5~]henicosa-1(20),2(21),16,18-tetraene-6,9,13-trione, Histone deacetylase 8, POTASSIUM ION, ... | | Authors: | Decroos, C, Christianson, D.W. | | Deposit date: | 2014-10-22 | | Release date: | 2015-04-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Variable Active Site Loop Conformations Accommodate the Binding of Macrocyclic Largazole Analogues to HDAC8.
Biochemistry, 54, 2015
|
|
4RLW
 
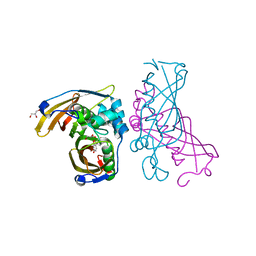 | | Crystal Structure of (3R)-hydroxyacyl-ACP dehydratase HadAB hetero-dimer from Mycobacterium tuberculosis complexed with Butein | | Descriptor: | (2E)-1-(2,4-dihydroxyphenyl)-3-(3,4-dihydroxyphenyl)prop-2-en-1-one, (3R)-hydroxyacyl-ACP dehydratase subunit HadA, (3R)-hydroxyacyl-ACP dehydratase subunit HadB, ... | | Authors: | Li, J, Dong, Y, Rao, Z.H. | | Deposit date: | 2014-10-18 | | Release date: | 2015-10-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.196 Å) | | Cite: | Molecular basis for the inhibition of beta-hydroxyacyl-ACP dehydratase HadAB complex from Mycobacterium tuberculosis by flavonoid inhibitors.
Protein Cell, 6, 2015
|
|
5KSJ
 
 | | Crystal structure of deoxygenated hemoglobin in complex with Sphingosine phosphate | | Descriptor: | (2S,3R,4E)-2-amino-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, Hemoglobin subunit alpha, Hemoglobin subunit beta, ... | | Authors: | Ahmed, M.H, Safo, M.K, Xia, Y. | | Deposit date: | 2016-07-08 | | Release date: | 2017-07-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and Functional Insight of Sphingosine 1-Phosphate-Mediated Pathogenic Metabolic Reprogramming in Sickle Cell Disease.
Sci Rep, 7, 2017
|
|
5L8W
 
 | | Structure of USP12-UB-PRG/UAF1 | | Descriptor: | GLYCEROL, Polyubiquitin-B, Ubiquitin carboxyl-terminal hydrolase 12, ... | | Authors: | Dharadhar, S, Sixma, T. | | Deposit date: | 2016-06-08 | | Release date: | 2016-09-28 | | Last modified: | 2016-11-30 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | A conserved two-step binding for the UAF1 regulator to the USP12 deubiquitinating enzyme.
J.Struct.Biol., 196, 2016
|
|
5KV9
 
 | | Crystal structure of a hPIV haemagglutinin-neuraminidase-inhibitor complex | | Descriptor: | 1,2-ETHANEDIOL, 2,6-anhydro-3,4,5-trideoxy-5-[(2-methylpropanoyl)amino]-4-(4-phenyl-1H-1,2,3-triazol-1-yl)-D-glycero-D-galacto-non-2-en onic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Dirr, L, El-Deeb, I.M, Chavas, L.M.G, Guillon, P, von Itzstein, M. | | Deposit date: | 2016-07-13 | | Release date: | 2017-07-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The impact of the butterfly effect on human parainfluenza virus haemagglutinin-neuraminidase inhibitor design.
Sci Rep, 7, 2017
|
|
4RME
 
 | | Crystal structure of human Retinoid X receptor alpha ligand binding domain complex with 9cUAB111 and coactivator peptide GRIP-1 | | Descriptor: | (2E,4E,6Z,8E)-3,7-dimethyl-8-[2-(3-methylbutyl)-3-(propan-2-yl)cyclohex-2-en-1-ylidene]octa-2,4,6-trienoic acid, Nuclear receptor coactivator 2, Retinoic acid receptor RXR-alpha | | Authors: | Xia, G, Muccio, D.D. | | Deposit date: | 2014-10-21 | | Release date: | 2015-09-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Conformationally Defined Rexinoids and Their Efficacy in the Prevention of Mammary Cancers.
J.Med.Chem., 58, 2015
|
|
4RSE
 
 | | Crystal structure of RPE65 in complex with MB-001 and palmitate | | Descriptor: | (1R)-3-amino-1-{3-[(2,6,6-trimethylcyclohex-1-en-1-yl)methoxy]phenyl}propan-1-ol, FE (II) ION, PALMITIC ACID, ... | | Authors: | Kiser, P.D, Shi, W, Palczewski, K. | | Deposit date: | 2014-11-07 | | Release date: | 2015-04-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Catalytic mechanism of a retinoid isomerase essential for vertebrate vision.
Nat.Chem.Biol., 11, 2015
|
|
4RN0
 
 | | Crystal structure of S39D HDAC8 in complex with a largazole analogue. | | Descriptor: | (5R,8S,11S)-5-methyl-8-(propan-2-yl)-11-[(1E)-4-sulfanylbut-1-en-1-yl]-3,17-dithia-7,10,14,19,20-pentaazatricyclo[14.2.1.1~2,5~]icosa-1(18),2(20),16(19)-triene-6,9,13-trione, GLYCEROL, Histone deacetylase 8, ... | | Authors: | Decroos, C, Christianson, D.W. | | Deposit date: | 2014-10-22 | | Release date: | 2015-04-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.761 Å) | | Cite: | Variable Active Site Loop Conformations Accommodate the Binding of Macrocyclic Largazole Analogues to HDAC8.
Biochemistry, 54, 2015
|
|
7N3U
 
 | | Crystal structure of human WEE1 kinase domain in complex with ZN-c3 | | Descriptor: | 1-[(7R)-7-ethyl-7-hydroxy-6,7-dihydro-5H-cyclopenta[b]pyridin-2-yl]-6-[4-(4-methylpiperazin-1-yl)anilino]-2-(prop-2-en-1-yl)-1,2-dihydro-3H-pyrazolo[3,4-d]pyrimidin-3-one, Wee1-like protein kinase | | Authors: | Lee, C.C. | | Deposit date: | 2021-06-02 | | Release date: | 2021-09-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Discovery of ZN-c3, a Highly Potent and Selective Wee1 Inhibitor Undergoing Evaluation in Clinical Trials for the Treatment of Cancer.
J.Med.Chem., 64, 2021
|
|
4RN1
 
 | | Crystal structure of S39D HDAC8 in complex with a largazole analogue. | | Descriptor: | (5R,8S,11S)-5-methyl-8-(propan-2-yl)-11-[(1E)-4-sulfanylbut-1-en-1-yl]-3-thia-7,10,14,20,21-pentaazatricyclo[14.3.1.1~2,5~]henicosa-1(20),2(21),16,18-tetraene-6,9,13-trione, GLYCEROL, Histone deacetylase 8, ... | | Authors: | Decroos, C, Christianson, D.W. | | Deposit date: | 2014-10-22 | | Release date: | 2015-04-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Variable Active Site Loop Conformations Accommodate the Binding of Macrocyclic Largazole Analogues to HDAC8.
Biochemistry, 54, 2015
|
|
