7WST
 
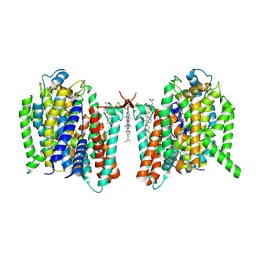 | | Cryo-EM structure of the barley Yellow stripe 1 transporter in complex with Fe(III)-DMA | | Descriptor: | (2~{S})-1-[(3~{S})-3-[[(3~{S})-3,4-bis(oxidanyl)-4-oxidanylidene-butyl]amino]-4-oxidanyl-4-oxidanylidene-butyl]azetidine-2-carboxylic acid, CHOLESTEROL HEMISUCCINATE, FE (III) ION, ... | | Authors: | Yamagata, A. | | Deposit date: | 2022-02-01 | | Release date: | 2022-11-30 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Uptake mechanism of iron-phytosiderophore from the soil based on the structure of yellow stripe transporter.
Nat Commun, 13, 2022
|
|
7ZQV
 
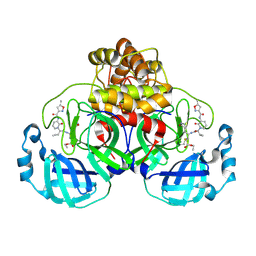 | | Structure of the SARS-CoV-2 main protease in complex with AG7404 | | Descriptor: | 3C-like proteinase nsp5, ethyl (4R)-4-({(2S)-2-[3-{[(5-methyl-1,2-oxazol-3-yl)carbonyl]amino}-2-oxopyridin-1(2H)-yl]pent-4-ynoyl}amino)-5-[(3S)-2-oxopyrrolidin-3-yl]pentanoate | | Authors: | Fabrega-Ferrer, M, Herrera-Morande, A, Perez-Saavedra, J, Coll, M. | | Deposit date: | 2022-05-03 | | Release date: | 2022-12-28 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structure and inhibition of SARS-CoV-1 and SARS-CoV-2 main proteases by oral antiviral compound AG7404.
Antiviral Res., 208, 2022
|
|
6SMH
 
 | | Cryo-electron microscopy structure of a RbcL-Raf1 supercomplex from Synechococcus elongatus PCC 7942 | | Descriptor: | Ribulose bisphosphate carboxylase large chain, Rubisco accumulation factor 1 (RAF1) peptide | | Authors: | Huang, F, Kong, W.-W, Sun, Y, Chen, T, Dykes, G.F, Jiang, Y.L, Liu, L.N. | | Deposit date: | 2019-08-21 | | Release date: | 2020-07-08 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Rubisco accumulation factor 1 (Raf1) plays essential roles in mediating Rubisco assembly and carboxysome biogenesis.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7AIF
 
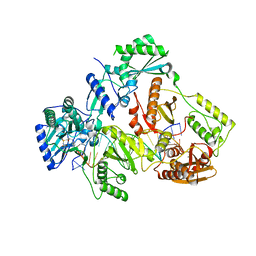 | | HIV-1 REVERSE TRANSCRIPTASE COMPLEX WITH DNA AND L-GLUTAMATE TENOFOVIR WITH BOUND MANGANESE | | Descriptor: | DNA (5'-D(*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*(DDG))-3'), DNA (5'-D(P*GP*GP*TP*CP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*G)-3'), Gag-Pol polyprotein, ... | | Authors: | Gu, W, Martinez, S.E, Nguyen, H, Xu, H, Herdewijn, P, de Jonghe, S, Das, K. | | Deposit date: | 2020-09-27 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Tenofovir-Amino Acid Conjugates Act as Polymerase Substrates-Implications for Avoiding Cellular Phosphorylation in the Discovery of Nucleotide Analogues.
J.Med.Chem., 64, 2021
|
|
6T2H
 
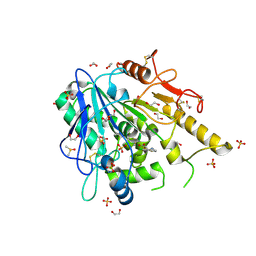 | | Furano[2,3-d]prymidine amides as Notum inhibitors | | Descriptor: | 1,2-ETHANEDIOL, 2-[[(4~{S})-5-chloranyl-6-methyl-1,2,3,4-tetrahydrothieno[2,3-d]pyrimidin-4-yl]sulfanyl]ethanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhao, Y, Jones, E.Y. | | Deposit date: | 2019-10-08 | | Release date: | 2020-01-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Scaffold-hopping identifies furano[2,3-d]pyrimidine amides as potent Notum inhibitors.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
5WN4
 
 | |
8A8Z
 
 | | Crystal structure of Danio rerio HDAC6 CD2 in complex with in situ enzymatically hydrolyzed DFMO-based ITF5924 | | Descriptor: | 4-[[4-[4-(imidazolidin-2-ylideneamino)phenyl]-1,2,3-triazol-1-yl]methyl]benzohydrazide, Histone deacetylase 6, POTASSIUM ION, ... | | Authors: | Zrubek, K, Sandrone, G, Cukier, C.D, Stevenazzi, A. | | Deposit date: | 2022-06-27 | | Release date: | 2023-01-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Difluoromethyl-1,3,4-oxadiazoles are slow-binding substrate analog inhibitors of histone deacetylase 6 with unprecedented isotype selectivity.
J.Biol.Chem., 299, 2022
|
|
7AID
 
 | | HIV-1 REVERSE TRANSCRIPTASE COMPLEX WITH DNA AND D-ASPARTATE TENOFOVIR | | Descriptor: | D-Aspartate Tenofovir, DNA (5'-D(*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*(DDG))-3'), DNA (5'-D(P*GP*GP*TP*CP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*G)-3'), ... | | Authors: | Gu, W, Martinez, S.E, Nguyen, H, Xu, H, Herdewijn, P, de Jonghe, S, Das, K. | | Deposit date: | 2020-09-26 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Tenofovir-Amino Acid Conjugates Act as Polymerase Substrates-Implications for Avoiding Cellular Phosphorylation in the Discovery of Nucleotide Analogues.
J.Med.Chem., 64, 2021
|
|
8AFR
 
 | | Pim1 in complex with 4-((6-hydroxybenzofuran-3-yl)methyl)benzoic acid and Pimtide | | Descriptor: | 4-((6-hydroxybenzofuran-3-yl)methyl)benzoic acid, Pimtide, Serine/threonine-protein kinase pim-1 | | Authors: | Hochban, P.M.M, Heine, A, Diederich, W.E. | | Deposit date: | 2022-07-18 | | Release date: | 2023-02-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Pose, duplicate, then elaborate: Steps towards increased affinity for inhibitors targeting the specificity surface of the Pim-1 kinase.
Eur.J.Med.Chem., 245, 2023
|
|
8KHU
 
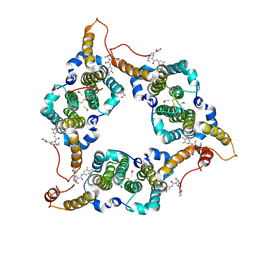 | | Hepatitis B virus core protein Y132A mutant in complex with THPP derivatives 48 | | Descriptor: | (6~{S},7~{R})-6,7-dimethyl-3-(2-oxidanylidenepyrrolidin-1-yl)-~{N}-[3,4,5-tris(fluoranyl)phenyl]-6,7-dihydro-4~{H}-pyrazolo[1,5-a]pyrazine-5-carboxamide, Capsid protein, GLYCEROL, ... | | Authors: | Zhou, Z, Xu, Z.H. | | Deposit date: | 2023-08-22 | | Release date: | 2023-10-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of 4,5,6,7-Tetrahydropyrazolo[1.5-a]pyrizine Derivatives as Core Protein Allosteric Modulators (CpAMs) for the Inhibition of Hepatitis B Virus.
J.Med.Chem., 66, 2023
|
|
6EQ2
 
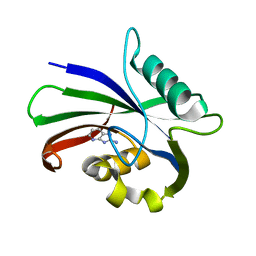 | | MTH1 in complex with fragment 6 | | Descriptor: | 1~{H}-imidazo[4,5-b]pyridin-2-amine, 7,8-dihydro-8-oxoguanine triphosphatase | | Authors: | Wiedmer, L, Sledz, P, Caflisch, A. | | Deposit date: | 2017-10-12 | | Release date: | 2018-10-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Ligand retargeting by binding site analogy.
Eur.J.Med.Chem., 175, 2019
|
|
8HA0
 
 | | Molecular recognition of two endogenous hormones by the human parathyroid hormone receptor-1 | | Descriptor: | CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhao, L, Xu, H.E, Yuan, Q. | | Deposit date: | 2022-10-26 | | Release date: | 2022-12-21 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.62 Å) | | Cite: | Molecular recognition of two endogenous hormones by the human parathyroid hormone receptor-1.
Acta Pharmacol.Sin., 44, 2023
|
|
6NC2
 
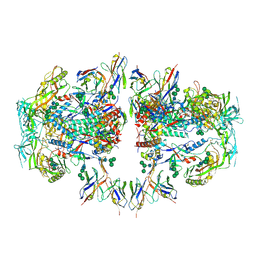 | | AMC011 v4.2 SOSIP Env trimer in complex with fusion peptide targeting antibody ACS202 fragment antigen binding | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, AMC011 v4.2 SOSIP gp120, ... | | Authors: | Cottrell, C.A, Ozorowski, G, Yuan, M, Copps, J, Wilson, I.A, Ward, A.B. | | Deposit date: | 2018-12-10 | | Release date: | 2019-06-19 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (5.2 Å) | | Cite: | Conformational Plasticity in the HIV-1 Fusion Peptide Facilitates Recognition by Broadly Neutralizing Antibodies.
Cell Host Microbe, 25, 2019
|
|
8HYG
 
 | |
7ZW5
 
 | | Crystal Structure of Unlinked NS2B-NS3 Protease from Zika Virus in Complex with Inhibitor MI-2259 | | Descriptor: | 1-[(8R,15S,18S)-15-(4-azanylbutyl)-18-(2-carbamimidamidoethyl)-4,7,14,17,20-pentakis(oxidanylidene)-3,6,13,16,19-pentazabicyclo[20.3.1]hexacosa-1(25),22(26),23-trien-8-yl]guanidine, Serine protease NS3, Serine protease subunit NS2B | | Authors: | Huber, S, Steinmetzer, T. | | Deposit date: | 2022-05-18 | | Release date: | 2023-05-31 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Synthesis and structural characterization of new macrocyclic inhibitors of the Zika virus NS2B-NS3 protease.
Arch Pharm, 2024
|
|
6E6V
 
 | | The N-terminal domain of PA endonuclease from the influenza H1N1 virus in complex with 3-hydroxy-6-methyl-4-oxo-1,4-dihydropyridine-2-carboxylic acid | | Descriptor: | 1,2-ETHANEDIOL, 3-hydroxy-6-methyl-4-oxo-1,4-dihydropyridine-2-carboxylic acid, MANGANESE (II) ION, ... | | Authors: | Dick, B.L, Morrison, C.N, Credille, C.V, Cohen, S.M. | | Deposit date: | 2018-07-25 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | SAR Exploration of Tight-Binding Inhibitors of Influenza Virus PA Endonuclease.
J.Med.Chem., 62, 2019
|
|
6EEU
 
 | | Structure of class II HMG-CoA reductase from Delftia acidovorans | | Descriptor: | 1,2-ETHANEDIOL, 3-hydroxy-3-methylglutaryl coenzyme A reductase, SULFATE ION | | Authors: | Ragwan, E.R, Arai, E, Kung, Y. | | Deposit date: | 2018-08-15 | | Release date: | 2018-09-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | New Crystallographic Snapshots of Large Domain Movements in Bacterial 3-Hydroxy-3-methylglutaryl Coenzyme A Reductase.
Biochemistry, 57, 2018
|
|
5MXX
 
 | | Crystal structure of human SR protein kinase 1 (SRPK1) in complex with compound 1 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5-methyl-~{N}-[2-(4-methylpiperazin-1-yl)-5-(trifluoromethyl)phenyl]furan-2-carboxamide, ... | | Authors: | Tallant, C, Redondo, C, Batson, J, Toop, H.D, Babaebi-Jadidib, R, Savitsky, P, Elkins, J.M, Newman, J.A, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bates, D.O, Morris, J.C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-01-25 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Development of Potent, Selective SRPK1 Inhibitors as Potential Topical Therapeutics for Neovascular Eye Disease.
ACS Chem. Biol., 12, 2017
|
|
8EPL
 
 | | Human R-type voltage-gated calcium channel Cav2.3 at 3.1 Angstrom resolution | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gao, S, Yao, X, Yan, N. | | Deposit date: | 2022-10-06 | | Release date: | 2022-12-14 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structures of the R-type human Ca v 2.3 channel reveal conformational crosstalk of the intracellular segments.
Nat Commun, 13, 2022
|
|
7B5V
 
 | | The carbohydrate binding module family 48 (CBM48) and carboxy-terminal carbohydrate esterase family 1 (CE1) domains of the multidomain esterase DmCE1B from Dysgonomonas mossii | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Carbohydrate Esterase family 1 protein with an N-terminal carbohydrate binding module family 48, ... | | Authors: | Mazurkewich, S, Kmezik, C, Branden, G, Larsbrink, J. | | Deposit date: | 2020-12-07 | | Release date: | 2021-03-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A polysaccharide utilization locus from the gut bacterium Dysgonomonas mossii encodes functionally distinct carbohydrate esterases.
J.Biol.Chem., 296, 2021
|
|
9JNF
 
 | | Crystal Structure of SME-1 E166A mutant in complex with Ertapenem | | Descriptor: | (2S,3R,4S)-4-({(3S,5S)-5-[(3-carboxyphenyl)carbamoyl]pyrrolidin-3-yl}sulfanyl)-2-[(1S,2R)-1-formyl-2-hydroxypropyl]-3-methyl-3,4-dihydro-2H-pyrrole-5-carboxylic acid, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Dhankhar, K, Hazra, S. | | Deposit date: | 2024-09-23 | | Release date: | 2025-10-01 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Crystal Structure of SME-1 E166A mutant in complex with Ertapenem
To Be Published
|
|
9BLZ
 
 | | State-1(phi motor) of full-length human dynein-1 in 5mM ATP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Cytoplasmic dynein 1 heavy chain 1, ... | | Authors: | Chai, P, Zhang, K. | | Deposit date: | 2024-05-02 | | Release date: | 2025-04-23 | | Last modified: | 2025-08-27 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | The mechanochemical cycle of reactive full-length human dynein 1.
Nat.Struct.Mol.Biol., 32, 2025
|
|
1K9W
 
 | | HIV-1(MAL) RNA Dimerization Initiation Site | | Descriptor: | HIV-1 DIS(Mal)UU RNA, MAGNESIUM ION | | Authors: | Ennifar, E, Walter, P, Ehresmann, B, Ehresmann, C, Dumas, P. | | Deposit date: | 2001-10-31 | | Release date: | 2001-11-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structures of coaxially stacked kissing complexes of the HIV-1 RNA dimerization initiation site.
Nat.Struct.Biol., 8, 2001
|
|
7TAZ
 
 | | Crystal structure of HIV-1 reverse transcriptase (RT) in complex with VM-1500A, a non-nucleoside RT inhibitor | | Descriptor: | 2-[4-bromo-3-(3-chloro-5-cyanophenoxy)-2-fluorophenyl]-N-(2-chloro-4-sulfamoylphenyl)acetamide, Reverse transcriptase p51, Reverse transcriptase/ribonuclease H | | Authors: | Snyder, A.A, Risener, C.J, Kirby, K.A, Sarafianos, S.G. | | Deposit date: | 2021-12-21 | | Release date: | 2023-04-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of HIV-1 reverse transcriptase (RT) in complex with VM-1500A, a non-nucleoside RT inhibitor
To Be Published
|
|
6EJB
 
 | |
