2H96
 
 | | Discovery of Potent, Highly Selective, and Orally Bioavailable Pyridine Carboxamide C-jun NH2-terminal Kinase Inhibitors | | Descriptor: | 5-CYANO-N-(2,5-DIMETHOXYBENZYL)-6-ETHOXYPYRIDINE-2-CARBOXAMIDE, C-jun-amino-terminal kinase-interacting protein 1, GLYCEROL, ... | | Authors: | Abad-Zapatero, C. | | Deposit date: | 2006-06-09 | | Release date: | 2006-07-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Discovery of potent, highly selective, and orally bioavailable pyridine carboxamide c-Jun NH2-terminal kinase inhibitors.
J.Med.Chem., 49, 2006
|
|
2GMX
 
 | | Selective Aminopyridine-Based C-Jun N-terminal Kinase inhibitors with cellular activity | | Descriptor: | C-jun-amino-terminal kinase-interacting protein 1, Mitogen-activated protein kinase 8, N-(4-AMINO-5-CYANO-6-ETHOXYPYRIDIN-2-YL)-2-(4-BROMO-2,5-DIMETHOXYPHENYL)ACETAMIDE, ... | | Authors: | Abad-Zapatero, C. | | Deposit date: | 2006-04-07 | | Release date: | 2006-06-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Aminopyridine-Based c-Jun N-Terminal Kinase Inhibitors with Cellular Activity and Minimal Cross-Kinase Activity.
J.Med.Chem., 49, 2006
|
|
4DEH
 
 | | Crystal structure of c-Met in complex with triazolopyridinone inhibitor 3 | | Descriptor: | 5-phenyl-3-(quinolin-6-ylmethyl)-3,5,6,7-tetrahydro-4H-[1,2,3]triazolo[4,5-c]pyridin-4-one, Hepatocyte growth factor receptor | | Authors: | Whittington, D.A, Bellon, S.F, Long, A.M. | | Deposit date: | 2012-01-20 | | Release date: | 2012-05-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery and optimization of a potent and selective triazolopyridinone series of c-Met inhibitors.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
6YYJ
 
 | | Crystal structure of native Phycocyanin from T. elongatus in spacegroup P21212 at 2.1 Angstroms | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-[5-[[(3~{R},4~{R})-3-ethyl-4-methyl-5-oxidanylidene-3,4-dihydropyrrol-2-yl]methyl]-2-[[5-[(~{Z})-(4-ethyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1~{H}-pyrrol-2-yl]methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, C-phycocyanin alpha chain, ... | | Authors: | Feiler, C.G, Falke, S, Sarrou, I. | | Deposit date: | 2020-05-05 | | Release date: | 2021-01-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | C-phycocyanin as a highly attractive model system in protein crystallography: unique crystallization properties and packing-diversity screening.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
1W2A
 
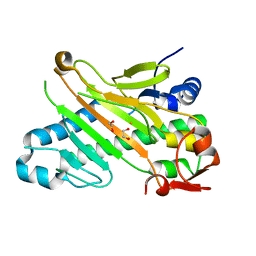 | | Deacetoxycephalosporin C synthase (with his-tag) complexed with Fe(II) and ethylene glycol | | Descriptor: | 1,2-ETHANEDIOL, DEACETOXYCEPHALOSPORIN C SYNTHASE, FE (III) ION | | Authors: | Oster, L.M, Terwisscha Van Scheltinga, A.C, Valegard, K, Mackenzie Hose, A, Dubus, A, Hajdu, J, Andersson, I. | | Deposit date: | 2004-07-01 | | Release date: | 2004-09-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Conformational Flexibility of the C Terminus with Implications for Substrate Binding and Catalysis Revealed in a New Crystal Form of Deacetoxycephalosporin C Synthase
J.Mol.Biol., 343, 2004
|
|
6QKN
 
 | | Structure of the azide-inhibited form of cytochrome c peroxidase from obligate human pathogenic bacterium Neisseria gonorrhoeae | | Descriptor: | AZIDE ION, CALCIUM ION, Cytochrome-c peroxidase, ... | | Authors: | Carvalho, A.L, Romao, M.J, Pauleta, S, Nobrega, C. | | Deposit date: | 2019-01-29 | | Release date: | 2020-02-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the mixed-valence, active form, of cytochrome c peroxidase from obligate human pathogenic bacterium Neisseria gonorrhoeae
To Be Published
|
|
1I52
 
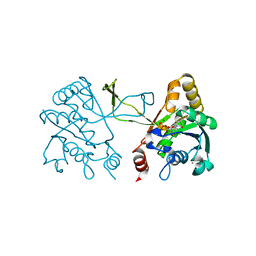 | | CRYSTAL STRUCTURE OF 4-DIPHOSPHOCYTIDYL-2-C-METHYLERYTHRITOL (CDP-ME) SYNTHASE (YGBP) INVOLVED IN MEVALONATE INDEPENDENT ISOPRENOID BIOSYNTHESIS | | Descriptor: | 4-DIPHOSPHOCYTIDYL-2-C-METHYLERYTHRITOL SYNTHASE, CALCIUM ION, CYTIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Richard, S.B, Bowman, M.E, Kwiatkowski, W, Kang, I, Chow, C, Lillo, M, Cane, D.E, Noel, J.P. | | Deposit date: | 2001-02-23 | | Release date: | 2001-07-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of 4-diphosphocytidyl-2-C- methylerythritol synthetase involved in mevalonate- independent isoprenoid biosynthesis.
Nat.Struct.Biol., 8, 2001
|
|
4XXL
 
 | |
8YT7
 
 | |
6GHD
 
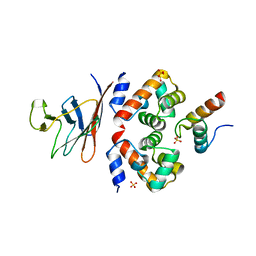 | | Structural analysis of the ternary complex between lamin A/C, BAF and emerin identifies an interface disrupted in autosomal recessive progeroid diseases | | Descriptor: | 1,2-ETHANEDIOL, Barrier-to-autointegration factor, Emerin, ... | | Authors: | Samson, C, Petitalot, A, Celli, F, Herrada, I, Ropars, V, Ledu, M.H, Nhiri, N, Arteni, A.A, Buendia, B, ZinnJustin, S. | | Deposit date: | 2018-05-07 | | Release date: | 2018-08-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural analysis of the ternary complex between lamin A/C, BAF and emerin identifies an interface disrupted in autosomal recessive progeroid diseases.
Nucleic Acids Res., 46, 2018
|
|
1I6E
 
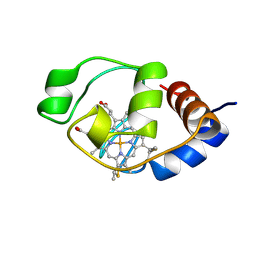 | | SOLUTION STRUCTURE OF THE FUNCTIONAL DOMAIN OF PARACOCCUS DENITRIFICANS CYTOCHROME C552 IN THE OXIDIZED STATE | | Descriptor: | CYTOCHROME C552, HEME C | | Authors: | Reincke, B, Perez, C, Pristovsek, P, Luecke, C, Ludwig, C, Loehr, F, Rogov, V.V, Ludwig, B, Rueterjans, H. | | Deposit date: | 2001-03-02 | | Release date: | 2001-10-17 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of the functional domain of Paracoccus denitrificans cytochrome c(552) in both redox states.
Biochemistry, 40, 2001
|
|
1I6D
 
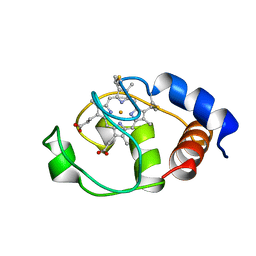 | | SOLUTION STRUCTURE OF THE FUNCTIONAL DOMAIN OF PARACOCCUS DENITRIFICANS CYTOCHROME C552 IN THE REDUCED STATE | | Descriptor: | CYTOCHROME C552, HEME C | | Authors: | Reincke, B, Perez, C, Pristovsek, P, Luecke, C, Ludwig, C, Loehr, F, Rogov, V.V, Ludwig, B, Rueterjans, H. | | Deposit date: | 2001-03-02 | | Release date: | 2001-10-17 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of the functional domain of Paracoccus denitrificans cytochrome c(552) in both redox states.
Biochemistry, 40, 2001
|
|
1HJG
 
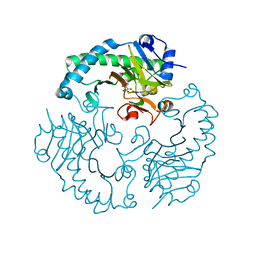 | | Alteration of the co-substrate selectivity of deacetoxycephalosporin C synthase: The role of arginine-258 | | Descriptor: | 3-METHYL-2-OXOBUTANOIC ACID, DEACETOXYCEPHALOSPORIN C SYNTHASE, FE (II) ION | | Authors: | Lee, H.J, Lloyd, M.D, Clifton, I.J, Harlos, K, Dubus, A, Baldwin, J.E, Frere, J.M, Schofield, C.J. | | Deposit date: | 2001-01-15 | | Release date: | 2001-06-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Alteration of the 2-Oxoacid Cosubstrate Selectivity in Deacetoxycephalosporin C Synthase: The Role of Arginine-258
J.Biol.Chem., 276, 2001
|
|
4WJY
 
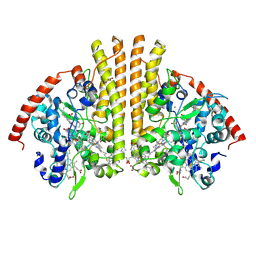 | | Esherichia coli nitrite reductase NrfA H264N | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Cytochrome c-552, ... | | Authors: | Clarke, T.A, Edwards, M.J, Lockwood, C.W.J. | | Deposit date: | 2014-10-01 | | Release date: | 2015-03-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Resolution of key roles for the distal pocket histidine in cytochrome C nitrite reductases.
J.Am.Chem.Soc., 137, 2015
|
|
8OHM
 
 | |
8FSJ
 
 | | Cryo-EM structure of engineered hepatitis C virus E1E2 ectodomain in complex with antibodies AR4A, HEPC74, and IGH520 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, AR4A heavy chain, ... | | Authors: | Metcalf, M.C, Ofek, G. | | Deposit date: | 2023-01-10 | | Release date: | 2023-07-19 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | Structure of engineered hepatitis C virus E1E2 ectodomain in complex with neutralizing antibodies.
Nat Commun, 14, 2023
|
|
8TAR
 
 | | APC/C-CDH1-UBE2C-Ubiquitin-CyclinB-NTD | | Descriptor: | Anaphase-promoting complex subunit 1, Anaphase-promoting complex subunit 10, Anaphase-promoting complex subunit 11, ... | | Authors: | Bodrug, T, Welsh, K.A, Bolhuis, D.L, Paulakonis, E, Martinez-Chacin, R.C, Liu, B, Pinkin, N, Bonacci, T, Cui, L, Xu, P, Roscow, O, Amann, S.J, Grishkovskaya, I, Emanuele, M.J, Harrison, J.S, Steimel, J.P, Hahn, K.M, Zhang, W, Zhong, E, Haselbach, D, Brown, N.G. | | Deposit date: | 2023-06-27 | | Release date: | 2023-09-27 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Time-resolved cryo-EM (TR-EM) analysis of substrate polyubiquitination by the RING E3 anaphase-promoting complex/cyclosome (APC/C).
Nat.Struct.Mol.Biol., 30, 2023
|
|
7B3Q
 
 | | Crystal structure of c-MET bound by compound 1 | | Descriptor: | 1-(phenylmethyl)-5~{H}-pyrrolo[3,2-c]pyridin-4-one, Hepatocyte growth factor receptor, SULFATE ION | | Authors: | Collie, G.W. | | Deposit date: | 2020-12-01 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Basis for Targeting the Folded P-Loop Conformation of c-MET.
Acs Med.Chem.Lett., 12, 2021
|
|
6PFV
 
 | | Structure of S. venezuelae RisG-WhiG-c-di-GMP complex: orthorhombic crystal form | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), AmfC protein, RNA polymerase sigma factor | | Authors: | Schumacher, M.A. | | Deposit date: | 2019-06-22 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | c-di-GMP Arms an Anti-sigma to Control Progression of Multicellular Differentiation in Streptomyces.
Mol.Cell, 77, 2020
|
|
6YQ8
 
 | | Crystal structure of native Phycocyanin from T. elongatus in spacegroup P63 at 1.8 Angstroms | | Descriptor: | 1,2-ETHANEDIOL, C-phycocyanin alpha chain, C-phycocyanin beta chain, ... | | Authors: | Feiler, C.G, Falke, S, Sarrou, I. | | Deposit date: | 2020-04-16 | | Release date: | 2021-01-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | C-phycocyanin as a highly attractive model system in protein crystallography: unique crystallization properties and packing-diversity screening.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
9GL7
 
 | | EGFR Exon20 insertion mutant NPG bound with (S)-3-((3-chloro-2-methoxyphenyl)amino)-2-(3-((tetrahydrofuran-2-yl)methoxy)pyridin-4-yl)-1,5,6,7-tetrahydro-4H-pyrrolo[3,2-c]pyridin-4-one | | Descriptor: | 3-[(3-chloranyl-2-methoxy-phenyl)amino]-2-[3-[[(2S)-oxolan-2-yl]methoxy]pyridin-4-yl]-1,5,6,7-tetrahydropyrrolo[3,2-c]pyridin-4-one, Epidermal growth factor receptor | | Authors: | Hilbert, B.J, Brooijmans, N, Milgram, B.C, Pagliarini, R.A. | | Deposit date: | 2024-08-27 | | Release date: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (1.878 Å) | | Cite: | EGFR Exon20 insertion mutant NPG bound with (S)-3-((3-chloro-2-methoxyphenyl)amino)-2-(3-((tetrahydrofuran-2-yl)methoxy)pyridin-4-yl)-1,5,6,7-tetrahydro-4H-pyrrolo[3,2-c]pyridin-4-one
To Be Published
|
|
4QO5
 
 | | Hypothetical multiheme protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, HEME C, ... | | Authors: | Rajendran, C. | | Deposit date: | 2014-06-19 | | Release date: | 2015-12-23 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.697 Å) | | Cite: | In meso crystal structure of a novel membrane-associated octaheme cytochrome c from the Crenarchaeon Ignicoccus hospitalis.
Febs J., 283, 2016
|
|
4RA5
 
 | | Human Protein Kinase C THETA IN COMPLEX WITH LIGAND COMPOUND 11a (6-[(1,3-Dimethyl-azetidin-3-yl)-methyl-amino]-4(R)-methyl-7-phenyl-2,10-dihydro-9-oxa-1,2,4a-triaza-phenanthren-3-one) | | Descriptor: | (1R)-9-[(1,3-dimethylazetidin-3-yl)(methyl)amino]-1-methyl-8-phenyl-3,5-dihydro[1,2,4]triazino[3,4-c][1,4]benzoxazin-2(1H)-one, 1,2-ETHANEDIOL, HUMAN PROTEIN KINASE C THETA, ... | | Authors: | Argiriadi, M.A, George, D.M. | | Deposit date: | 2014-09-09 | | Release date: | 2014-10-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Optimized Protein Kinase C theta (PKC theta ) Inhibitors Reveal Only Modest Anti-inflammatory Efficacy in a Rodent Model of Arthritis.
J.Med.Chem., 58, 2015
|
|
5KE0
 
 | | Discovery of 1-1H-Pyrazolo 4,3-c pyridine-6-yl urea Inhibitors of Extracellular Signal Regulated Kinase ERK for the Treatment of Cancers | | Descriptor: | 1-[3-(2-methylpyridin-4-yl)-1~{H}-pyrazolo[4,3-c]pyridin-6-yl]-3-(phenylmethyl)urea, Mitogen-activated protein kinase 1, SULFATE ION | | Authors: | Hruza, A, Lim, J. | | Deposit date: | 2016-06-09 | | Release date: | 2016-07-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Discovery of 1-(1H-Pyrazolo[4,3-c]pyridin-6-yl)urea Inhibitors of Extracellular Signal-Regulated Kinase (ERK) for the Treatment of Cancers.
J.Med.Chem., 59, 2016
|
|
5NGX
 
 | | The 1.06 A resolution structure of the L16G mutant of ferric cytochrome c prime from Alcaligenes xylosoxidans, complexed with nitrite | | Descriptor: | Cytochrome c', GLYCEROL, HEME C, ... | | Authors: | Strange, R, Hough, M, Kekelli, D, Horrell, S, Moreno Chicano, T. | | Deposit date: | 2017-03-20 | | Release date: | 2018-05-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Distinguishing Nitro vs Nitrito Coordination in Cytochrome c' Using Vibrational Spectroscopy and Density Functional Theory.
Inorg.Chem., 56, 2017
|
|
