4XK9
 
 | | Crystal structure of A-AChBP in complex with pinnatoxin G | | Descriptor: | CHLORIDE ION, Pinnatoxin G, Soluble acetylcholine receptor | | Authors: | Bourne, Y, Sulzenbacher, G, Marchot, P. | | Deposit date: | 2015-01-10 | | Release date: | 2015-06-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Marine Macrocyclic Imines, Pinnatoxins A and G: Structural Determinants and Functional Properties to Distinguish Neuronal alpha 7 from Muscle alpha 12 beta gamma delta nAChRs.
Structure, 23, 2015
|
|
4XL1
 
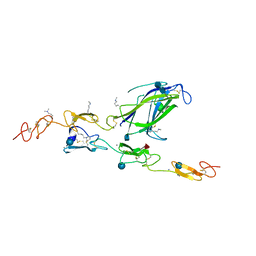 | | Complex of Notch1 (EGF11-13) bound to Delta-like 4 (N-EGF1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Luca, V.C, Jude, K.M, Garcia, K.C. | | Deposit date: | 2015-01-13 | | Release date: | 2015-03-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural biology. Structural basis for Notch1 engagement of Delta-like 4.
Science, 347, 2015
|
|
4XLW
 
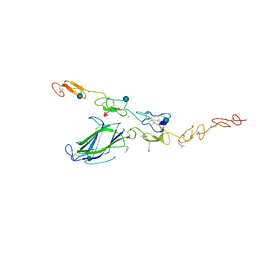 | | Complex of Notch1 (EGF11-13) bound to Delta-like 4 (N-EGF2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Delta-like protein, ... | | Authors: | Luca, V.C, Jude, K.M, Garcia, K.C. | | Deposit date: | 2015-01-13 | | Release date: | 2015-03-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.39 Å) | | Cite: | Structural biology. Structural basis for Notch1 engagement of Delta-like 4.
Science, 347, 2015
|
|
5EQ6
 
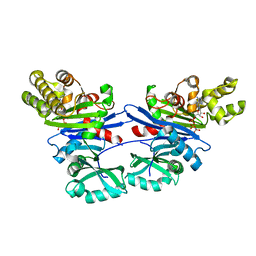 | | Pseudomonas aeruginosa PilM bound to AMP-PNP | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Type 4 fimbrial biogenesis protein PilM | | Authors: | McCallum, M, Tammam, S, Robinson, H, Shah, M, Calmettes, C, Moraes, T, Burrows, L.L, Howell, P.L. | | Deposit date: | 2015-11-12 | | Release date: | 2016-04-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | PilN Binding Modulates the Structure and Binding Partners of the Pseudomonas aeruginosa Type IVa Pilus Protein PilM.
J.Biol.Chem., 291, 2016
|
|
3NO0
 
 | |
3NY1
 
 | | Structure of the ubr-box of the UBR1 ubiquitin ligase | | Descriptor: | E3 ubiquitin-protein ligase UBR1, ZINC ION | | Authors: | Matta-Camacho, E, Kozlov, G, Li, F, Gehring, K. | | Deposit date: | 2010-07-14 | | Release date: | 2010-08-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.085 Å) | | Cite: | Structural basis of substrate recognition and specificity in the N-end rule pathway.
Nat.Struct.Mol.Biol., 17, 2010
|
|
5EOX
 
 | | Pseudomonas aeruginosa PilM bound to ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Type 4 fimbrial biogenesis protein PilM | | Authors: | McCallum, M, Tammam, S, Robinson, H, Shah, M, Calmettes, C, Moraes, T, Burrows, L.L, Howell, P.L. | | Deposit date: | 2015-11-10 | | Release date: | 2016-04-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | PilN Binding Modulates the Structure and Binding Partners of the Pseudomonas aeruginosa Type IVa Pilus Protein PilM.
J.Biol.Chem., 291, 2016
|
|
3EAX
 
 | | Crystal structure PTP1B complex with small molecule compound LZP-6 | | Descriptor: | 4,4'-piperazine-1,4-diylbis{1-[3-(benzyloxy)phenyl]-4-oxobutane-1,3-dione}, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Zhang, Z.-Y, Liu, S, Zhang, L.-F, Yu, X, Xue, T, Gunawan, A.M, Long, Y.-Q. | | Deposit date: | 2008-08-26 | | Release date: | 2009-07-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Targeting inactive enzyme conformation: aryl diketoacid derivatives as a new class of PTP1B inhibitors.
J.Am.Chem.Soc., 130, 2008
|
|
3O32
 
 | | Crystal Structure of 4-Chlorocatechol Dioxygenase from Rhodococcus opacus 1CP in complex with 3,5-dichlorocatechol | | Descriptor: | (2R)-3-(PHOSPHONOOXY)-2-(TETRADECANOYLOXY)PROPYL PALMITATE, 3,5-dichlorobenzene-1,2-diol, Chlorocatechol 1,2-dioxygenase, ... | | Authors: | Ferraroni, M, Briganti, F, Kolomytseva, M, Golovleva, L. | | Deposit date: | 2010-07-23 | | Release date: | 2011-08-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | X-ray structures of 4-chlorocatechol 1,2-dioxygenase adducts with substituted catechols: new perspectives in the molecular basis of intradiol ring cleaving dioxygenases specificity.
J. Struct. Biol., 181, 2013
|
|
3O5U
 
 | | Crystal Structure of 4-Chlorocatechol Dioxygenase from Rhodococcus opacus 1CP in complex with protocatechuate | | Descriptor: | (2R)-3-(PHOSPHONOOXY)-2-(TETRADECANOYLOXY)PROPYL PALMITATE, 3,4-DIHYDROXYBENZOIC ACID, CHLORIDE ION, ... | | Authors: | Ferraroni, M, Briganti, F, Kolomitseva, M, Golovleva, L. | | Deposit date: | 2010-07-28 | | Release date: | 2011-08-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | X-ray structures of 4-chlorocatechol 1,2-dioxygenase adducts with substituted catechols: new perspectives in the molecular basis of intradiol ring cleaving dioxygenases specificity.
J. Struct. Biol., 181, 2013
|
|
2M1W
 
 | | TICAM-2 TIR domain | | Descriptor: | TIR domain-containing adapter molecule 2 | | Authors: | Enokizono, Y, Kumeta, H, Funami, K, Horiuchi, M, Sarmiento, J, Yamashita, K, Standley, D.M, Matsumoto, M, Seya, T, Inagaki, F. | | Deposit date: | 2012-12-07 | | Release date: | 2014-01-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structures and interface mapping of the TIR domain-containing adaptor molecules involved in interferon signaling.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4EQU
 
 | | Human STK-10 (LOK) kinase domain in DFG-out conformation with inhibitor DSA-7 | | Descriptor: | CALCIUM ION, N-{3-[(3-{4-[(4-methoxyphenyl)amino]-1,3,5-triazin-2-yl}pyridin-2-yl)amino]-4-methylphenyl}-3-(trifluoromethyl)benzamide, PENTAETHYLENE GLYCOL, ... | | Authors: | Merritt, E.A, Larson, E.T. | | Deposit date: | 2012-04-19 | | Release date: | 2012-11-07 | | Last modified: | 2012-12-19 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Affinity-Based Probes Based on Type II Kinase Inhibitors.
J.Am.Chem.Soc., 134, 2012
|
|
6ZWT
 
 | | Crystal structure of DNA-binding domain of OmpR of two-component system of Acinetobacter baumannii | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, SULFATE ION, ... | | Authors: | Narwal, M, Lucchini, V, Trebosc, V, Gartenmann, S, Pieren, M, Kemmer, C, Kammerer, R.A. | | Deposit date: | 2020-07-28 | | Release date: | 2021-08-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Targeting virulence regulation to disarm Acinetobacter baumannii pathogenesis.
Virulence, 13, 2022
|
|
5XTZ
 
 | | Crystal structure of GAS41 YEATS bound to H3K27ac peptide | | Descriptor: | ACETATE ION, THR-LYS-ALA-ALA-ARG-ALY-SER-ALA-PRO-ALA, YEATS domain-containing protein 4 | | Authors: | Li, H.T, Zhao, D. | | Deposit date: | 2017-06-21 | | Release date: | 2018-06-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.105 Å) | | Cite: | Gas41 links histone acetylation to H2A.Z deposition and maintenance of embryonic stem cell identity.
Cell Discov, 4, 2018
|
|
2M6T
 
 | | NMR solution structure ensemble of 3-4D mutant domain 11 IGF2R | | Descriptor: | Insulin-like growth factor 2 receptor variant | | Authors: | Strickland, M, Williams, C, Richards, E, Minnall, L, Crump, M.P, Frago, S, Hughes, J, Garner, L, Hoppe, H, Rezgui, D, Zaccheo, O.J, Prince, S.N, Hassan, A.B, Whittaker, S. | | Deposit date: | 2013-04-09 | | Release date: | 2014-10-15 | | Last modified: | 2016-06-01 | | Method: | SOLUTION NMR | | Cite: | Functional evolution of IGF2:IGF2R domain 11 binding generates novel structural interactions and a specific IGF2 antagonist.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
2LWW
 
 | | NMR structure of RelA-TAD/CBP-TAZ1 complex | | Descriptor: | CREB-binding protein, Nuclear transcription factor RelA, ZINC ION | | Authors: | Mukherjee, S.P, Ghosh, G, Wright, P.E. | | Deposit date: | 2012-08-07 | | Release date: | 2013-09-11 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Analysis of the RelA:CBP/p300 Interaction Reveals Its Involvement in NF-kappa B-Driven Transcription.
Plos Biol., 11, 2013
|
|
4Y21
 
 | | Crystal Structure of Munc13-1 MUN domain | | Descriptor: | Protein unc-13 homolog A | | Authors: | Yang, X.Y, Wang, S, Sheng, Y, Zhang, M, Zou, W.J, Wu, L.J, Kang, L.J, Rizo, J, Zhang, R.G, Xu, T, Ma, C. | | Deposit date: | 2015-02-09 | | Release date: | 2015-06-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Syntaxin opening by the MUN domain underlies the function of Munc13 in synaptic-vesicle priming.
Nat.Struct.Mol.Biol., 22, 2015
|
|
4E96
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with the inhibitor PFi-1 | | Descriptor: | 1,2-ETHANEDIOL, 2-methoxy-N-(3-methyl-2-oxo-1,2,3,4-tetrahydroquinazolin-6-yl)benzenesulfonamide, Bromodomain-containing protein 4, ... | | Authors: | Filippakopoulos, P, Picaud, S, Felletar, I, Fedorov, O, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Fish, P, Bunnage, M, Owen, D, Knapp, S, Cook, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-03-20 | | Release date: | 2012-04-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Identification of a chemical probe for bromo and extra C-terminal bromodomain inhibition through optimization of a fragment-derived hit.
J.Med.Chem., 55, 2012
|
|
3EB5
 
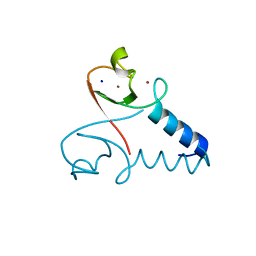 | | Structure of the cIAP2 RING domain | | Descriptor: | Baculoviral IAP repeat-containing protein 3, SODIUM ION, ZINC ION | | Authors: | Mace, P.D, Linke, K, Smith, C.A, Day, C.L. | | Deposit date: | 2008-08-27 | | Release date: | 2008-09-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of the cIAP2 RING domain reveal conformational changes associated with ubiquitin-conjugating enzyme (E2) recruitment.
J.Biol.Chem., 283, 2008
|
|
2LZ6
 
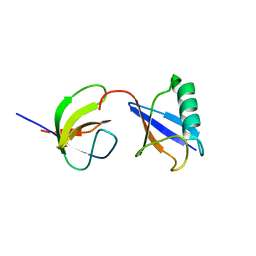 | | Distinct ubiquitin binding modes exhibited by sh3 domains: molecular determinants and functional implications | | Descriptor: | CD2-associated protein, Ubiquitin | | Authors: | Ortega-Roldan, J, Azuaga, A, Blackledge, M, Van Nuland, N. | | Deposit date: | 2012-09-24 | | Release date: | 2013-10-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Distinct Ubiquitin Binding Modes Exhibited by SH3 Domains: Molecular Determinants and Functional Implications.
Plos One, 8, 2013
|
|
4Y0J
 
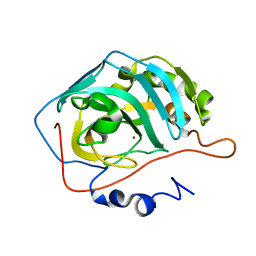 | |
3ODT
 
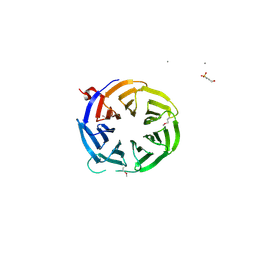 | | Crystal structure of WD40 beta propeller domain of Doa1 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, Protein DOA1 | | Authors: | Pashkova, N, Gakhar, L, Winistorfer, S.C, Yu, L, Ramaswamy, S, Piper, R.C. | | Deposit date: | 2010-08-11 | | Release date: | 2010-12-01 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | WD40 Repeat Propellers Define a Ubiquitin-Binding Domain that Regulates Turnover of F Box Proteins.
Mol.Cell, 40, 2010
|
|
3O6J
 
 | | Crystal Structure of 4-Chlorocatechol Dioxygenase from Rhodococcus opacus 1CP in complex with hydroxyquinol | | Descriptor: | (2R)-3-(PHOSPHONOOXY)-2-(TETRADECANOYLOXY)PROPYL PALMITATE, CHLORIDE ION, Chlorocatechol 1,2-dioxygenase, ... | | Authors: | Ferraroni, M, Briganti, F, Kolomitseva, M, Golovleva, L. | | Deposit date: | 2010-07-29 | | Release date: | 2011-08-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | X-ray structures of 4-chlorocatechol 1,2-dioxygenase adducts with substituted catechols: new perspectives in the molecular basis of intradiol ring cleaving dioxygenases specificity.
J. Struct. Biol., 181, 2013
|
|
5Y28
 
 | | Crystal structure of H. pylori HtrA with PDZ2 deletion | | Descriptor: | 1,2-ETHANEDIOL, Periplasmic serine endoprotease DegP-like, UNK-UNK-UNK-UNK | | Authors: | Zhang, Z, Huang, Q, Tao, X. | | Deposit date: | 2017-07-24 | | Release date: | 2018-08-15 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.08606815 Å) | | Cite: | The unique trimeric assembly of the virulence factor HtrA fromHelicobacter pylorioccurs via N-terminal domain swapping.
J.Biol.Chem., 294, 2019
|
|
4XHE
 
 | | Crystal Structure of A-AChBP in complex with pinnatoxin A | | Descriptor: | CALCIUM ION, CHLORIDE ION, Pinnatoxin A, ... | | Authors: | Bourne, Y, Sulzenbacher, G, Marchot, P. | | Deposit date: | 2015-01-05 | | Release date: | 2015-06-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Marine Macrocyclic Imines, Pinnatoxins A and G: Structural Determinants and Functional Properties to Distinguish Neuronal alpha 7 from Muscle alpha 12 beta gamma delta nAChRs.
Structure, 23, 2015
|
|
