4DUM
 
 | | Co-crystal structure of eIF4E with inhibitor | | Descriptor: | (4-{7-[2-(4-chlorophenoxy)ethyl]-2-(methylamino)-6-oxo-6,7-dihydro-1H-purin-8-yl}phenyl)phosphonic acid, 1,2-ETHANEDIOL, Eukaryotic translation initiation factor 4E | | Authors: | Min, X, Johnstone, S, Walker, N, Wang, Z. | | Deposit date: | 2012-02-22 | | Release date: | 2012-04-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structure-Guided Design, Synthesis, and Evaluation of Guanine-Derived Inhibitors of the eIF4E mRNA-Cap Interaction.
J.Med.Chem., 55, 2012
|
|
6SHN
 
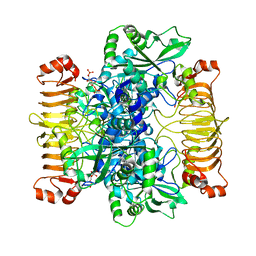 | | Escherichia coli AGPase in complex with FBP. Symmetry C1 | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, Glucose-1-phosphate adenylyltransferase | | Authors: | Cifuente, J.O, Comino, N, D'Angelo, C, Marina, A, Gil-Carton, D, Albesa-Jove, D, Guerin, M.E. | | Deposit date: | 2019-08-07 | | Release date: | 2020-02-05 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The allosteric control mechanism of bacterial glycogen biosynthesis disclosed by cryoEM.
Curr Res Struct Biol, 2, 2020
|
|
4DGV
 
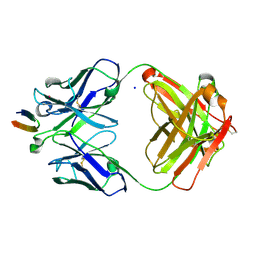 | | Structure of the Hepatitis C virus envelope glycoprotein E2 antigenic region 412-423 bound to the broadly neutralizing antibody HCV1, P2(1) form | | Descriptor: | E2 peptide, HCV1 Heavy Chain, HCV1 Light Chain, ... | | Authors: | Kong, L, Wilson, I.A, Law, M. | | Deposit date: | 2012-01-26 | | Release date: | 2012-05-23 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.805 Å) | | Cite: | Structural basis of hepatitis C virus neutralization by broadly neutralizing antibody HCV1.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
8P0I
 
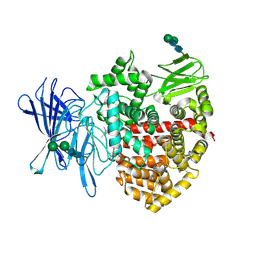 | | Crystal structure of the open conformation of insulin-regulated aminopeptidase in complex with a small-MW inhibitor | | Descriptor: | 2-[2-(3,5-dimethylpyrazol-1-yl)-6-(4-methoxyphenyl)pyrimidin-4-yl]oxy-N-methyl-N-[(2-methylpyridin-3-yl)methyl]ethanamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Mpakali, A, Giastas, P, Stratikos, E. | | Deposit date: | 2023-05-10 | | Release date: | 2024-09-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Stabilization of the open conformation omicron f insulin-regulated aminopeptidase by a novel substrate-selective small-molecule inhibitor.
Protein Sci., 33, 2024
|
|
2XGK
 
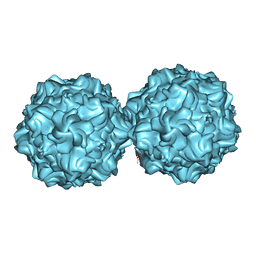 | | Virus like particle of L172W mutant of Minute Virus of Mice - the immunosuppressive strain | | Descriptor: | COAT PROTEIN VP2 | | Authors: | Plevka, P, Hafenstein, S, Tattersall, P, Cotmore, S, Farr, G, D'Abramo, A, Rossmann, M.G. | | Deposit date: | 2010-06-04 | | Release date: | 2011-04-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Structure of a Packaging-Defective Mutant of Minute Virus of Mice Indicates that the Genome is Packaged Via a Pore at a 5-Fold Axis.
J.Virol., 85, 2011
|
|
8P8B
 
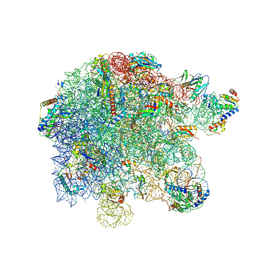 | | Mycoplasma pneumoniae large ribosomal subunit in chloramphenicol-treated cells | | Descriptor: | 1,4-DIAMINOBUTANE, 16S ribosomal RNA, 23S ribosomal RNA, ... | | Authors: | Schacherl, M, Xue, L, Spahn, C.M.T, Mahamid, J. | | Deposit date: | 2023-05-31 | | Release date: | 2024-11-20 | | Last modified: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural insights into context-dependent inhibitory mechanisms of chloramphenicol in cells.
Nat.Struct.Mol.Biol., 32, 2025
|
|
4H38
 
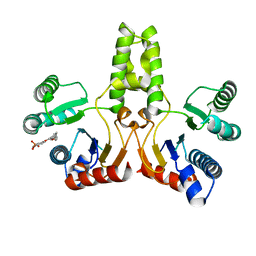 | |
8Q7R
 
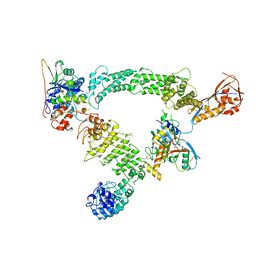 | | Ubiquitin ligation to substrate by a cullin-RING E3 ligase & Cdc34: NEDD8-CUL2-RBX1-ELOB/C-FEM1C with trapped UBE2R2~donor UB-Sil1 peptide | | Descriptor: | 5-azanyl-1-oxidanyl-pentan-2-one, Cullin-2, E3 ubiquitin-protein ligase RBX1, ... | | Authors: | Liwocha, J, Prabu, J.R, Kleiger, G, Schulman, B.A. | | Deposit date: | 2023-08-16 | | Release date: | 2024-02-21 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.71 Å) | | Cite: | Cullin-RING ligases employ geometrically optimized catalytic partners for substrate targeting.
Mol.Cell, 84, 2024
|
|
4H3C
 
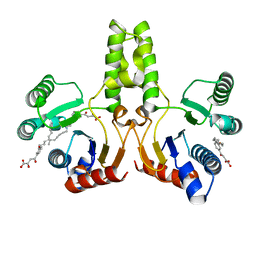 | |
2YJ1
 
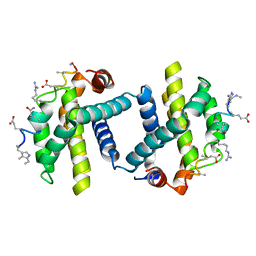 | | Puma BH3 foldamer in complex with Bcl-xL | | Descriptor: | ALPHA-BETA-PUMA BH3 FOLDAMER, BCL-2-LIKE PROTEIN 1 | | Authors: | Lee, E.F, Smith, B.J, Horne, W.S, Mayer, K.N, Evangelista, M, Colman, P.M, Gellman, S.H, Fairlie, W.D. | | Deposit date: | 2011-05-18 | | Release date: | 2011-10-12 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structural Basis of Bcl-Xl Recognition by a Bh3-Mimetic Alpha-Beta-Peptide Generated Via Sequence-Based Design
Chembiochem, 12, 2011
|
|
4DI5
 
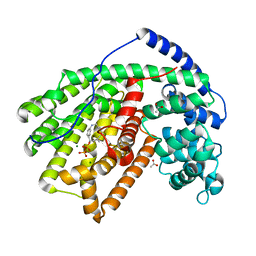 | |
5IK6
 
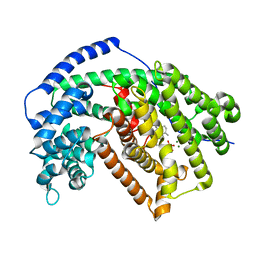 | | Tobacco 5-epi-aristolochene synthase with germacrene A and PPi | | Descriptor: | 5-epi-aristolochene synthase, DIPHOSPHATE, GERMACRENE A, ... | | Authors: | Koo, H.J, Xu, Y, Louie, G.V, Bowman, M, Noel, J.P. | | Deposit date: | 2016-03-03 | | Release date: | 2016-10-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Biosynthetic potential of sesquiterpene synthases: product profiles of Egyptian Henbane premnaspirodiene synthase and related mutants.
J.Antibiot., 69, 2016
|
|
6AKO
 
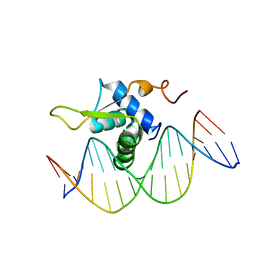 | | Crystal Structure of FOXC2 DBD Bound to DBE2 DNA | | Descriptor: | DNA (5'-D(CP*AP*AP*AP*AP*TP*GP*TP*AP*AP*AP*CP*AP*AP*GP*A)-3'), DNA (5'-D(TP*CP*TP*TP*GP*TP*TP*TP*AP*CP*AP*TP*TP*TP*TP*G)-3'), Forkhead box protein C2, ... | | Authors: | Chen, X, Wei, H, Li, J, Liang, X, Dai, S, Jiang, L, Guo, M, Chen, Y. | | Deposit date: | 2018-09-03 | | Release date: | 2019-02-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.396 Å) | | Cite: | Structural basis for DNA recognition by FOXC2.
Nucleic Acids Res., 47, 2019
|
|
4H2H
 
 | | Crystal structure of an enolase (mandalate racemase subgroup, target EFI-502101) from Pelagibaca bermudensis htcc2601, with bound mg and l-4-hydroxyproline betaine (betonicine) | | Descriptor: | (2S,4R)-4-hydroxy-1,1-dimethylpyrrolidinium-2-carboxylate, (4S)-2-METHYL-2,4-PENTANEDIOL, IODIDE ION, ... | | Authors: | Vetting, M.W, Morisco, L.L, Wasserman, S.R, Sojitra, S, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-09-12 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of new enzymes and metabolic pathways by using structure and genome context.
Nature, 502, 2013
|
|
4H03
 
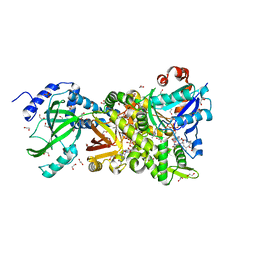 | | Crystal structure of NAD+-Ia-actin complex | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, Actin, ... | | Authors: | Tsurumura, T, Oda, M, Nagahama, M, Tsuge, H. | | Deposit date: | 2012-09-07 | | Release date: | 2013-02-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Arginine ADP-ribosylation mechanism based on structural snapshots of iota-toxin and actin complex
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
5IQR
 
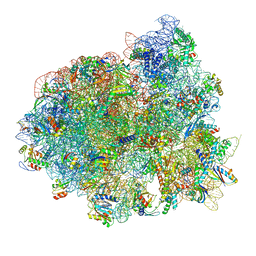 | | Structure of RelA bound to the 70S ribosome | | Descriptor: | 30S ribosomal protein S10, 30S ribosomal protein S11, 30S ribosomal protein S12, ... | | Authors: | Brown, A, Fernandez, I.S, Gordiyenko, Y, Ramakrishnan, V. | | Deposit date: | 2016-03-11 | | Release date: | 2016-05-04 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Ribosome-dependent activation of stringent control.
Nature, 534, 2016
|
|
4DOJ
 
 | | Crystal structure of BetP in outward-facing conformation | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, CHLORIDE ION, CHOLINE ION, ... | | Authors: | Perez, C, Ziegler, C. | | Deposit date: | 2012-02-09 | | Release date: | 2012-06-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Alternating-access mechanism in conformationally asymmetric trimers of the betaine transporter BetP.
Nature, 490, 2012
|
|
8QSO
 
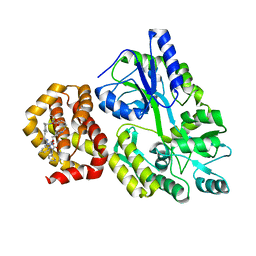 | | Crystal structure of human Mcl-1 in complex with compound 1 | | Descriptor: | (13S,16R,19S)-16-benzyl-43-ethoxy-N-methyl-7,11,14,17-tetraoxo-13-phenyl-5-oxa-2,8,12,15,18-pentaaza-1(1,4),4(1,2)-dibenzena-9(1,4)-cyclohexanacycloicosaphane-19-carboxamide, Maltose/maltodextrin-binding periplasmic protein,Induced myeloid leukemia cell differentiation protein Mcl-1, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Hekking, K.F.W, Gremmen, S, Maroto, S, Keefe, A.D, Zhang, Y. | | Deposit date: | 2023-10-10 | | Release date: | 2024-02-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.106 Å) | | Cite: | Development of Potent Mcl-1 Inhibitors: Structural Investigations on Macrocycles Originating from a DNA-Encoded Chemical Library Screen.
J.Med.Chem., 67, 2024
|
|
4H8E
 
 | |
8QY4
 
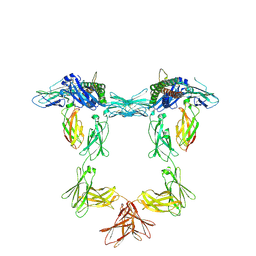 | | Structure of interleukin 11 (gp130 P496L mutant). | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-11, ... | | Authors: | Gardner, S, Bubeck, D, Jin, Y. | | Deposit date: | 2023-10-25 | | Release date: | 2024-02-21 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Structural insights into IL-11-mediated signalling and human IL6ST variant-associated immunodeficiency.
Nat Commun, 15, 2024
|
|
8QY5
 
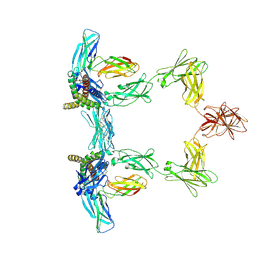 | | Structure of interleukin 6. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-6, ... | | Authors: | Gardner, S, Bubeck, D, Jin, Y. | | Deposit date: | 2023-10-25 | | Release date: | 2024-02-21 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into IL-11-mediated signalling and human IL6ST variant-associated immunodeficiency.
Nat Commun, 15, 2024
|
|
4F5C
 
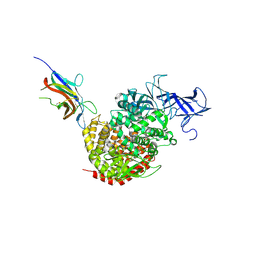 | | Crystal structure of the spike receptor binding domain of a porcine respiratory coronavirus in complex with the pig aminopeptidase N ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Aminopeptidase N, ... | | Authors: | Santiago, C, Reguera, J, Gaurav, M, Ordono, D, Enjuanes, L, Casasnovas, J.M. | | Deposit date: | 2012-05-13 | | Release date: | 2012-08-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural bases of coronavirus attachment to host aminopeptidase N and its inhibition by neutralizing antibodies.
Plos Pathog., 8, 2012
|
|
6SHJ
 
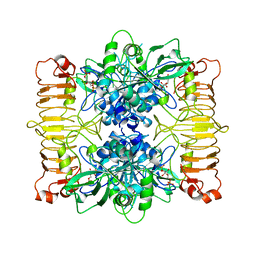 | | Escherichia coli AGPase in complex with FBP. Symmetry applied C2 | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, Glucose-1-phosphate adenylyltransferase | | Authors: | Cifuente, J.O, Comino, N, D'Angelo, C, Marina, A, Gil-Carton, D, Albesa-Jove, D, Guerin, M.E. | | Deposit date: | 2019-08-07 | | Release date: | 2020-02-05 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The allosteric control mechanism of bacterial glycogen biosynthesis disclosed by cryoEM.
Curr Res Struct Biol, 2, 2020
|
|
6SPF
 
 | | Pseudomonas aeruginosa 70s ribosome from an aminoglycoside resistant clinical isolate | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Halfon, Y, Jimenez-Fernande, A, La Ros, R, Espinos, R, Krogh Johansen, H, Matzov, D, Eyal, Z, Bashan, A, Zimmerman, E, Belousoff, M, Molin, S, Yonath, A. | | Deposit date: | 2019-09-01 | | Release date: | 2019-10-23 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Structure ofPseudomonas aeruginosaribosomes from an aminoglycoside-resistant clinical isolate.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
8QY6
 
 | | Structure of interleukin 6 (gp130 P496L mutant). | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-6, ... | | Authors: | Gardner, S, Bubeck, D, Jin, Y. | | Deposit date: | 2023-10-25 | | Release date: | 2024-02-21 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Structural insights into IL-11-mediated signalling and human IL6ST variant-associated immunodeficiency.
Nat Commun, 15, 2024
|
|
