8GM4
 
 | | Functional construct of the Eukaryotic elongation factor 2 kinase bound to an ATP-competitive inhibitor | | Descriptor: | 7-amino-1-cyclopropyl-3-ethyl-2,4-dioxo-1,2,3,4-tetrahydropyrido[2,3-d]pyrimidine-6-carboxamide, ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, ... | | Authors: | Piserchio, A, Isiorho, E.A, Dalby, K.N, Ghose, R. | | Deposit date: | 2023-03-24 | | Release date: | 2023-05-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structure of the complex between calmodulin and a functional construct of eukaryotic elongation factor 2 kinase bound to an ATP-competitive inhibitor.
J.Biol.Chem., 299, 2023
|
|
6XZP
 
 | | Influenza C virus polymerase in complex with chicken ANP32A - Subclass 4 | | Descriptor: | LRRcap domain-containing protein, Polymerase acidic protein, Polymerase basic protein 2, ... | | Authors: | Carrique, L, Keown, J.R, Fan, H, Grimes, J.M, Fodor, E. | | Deposit date: | 2020-02-05 | | Release date: | 2020-11-25 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Host ANP32A mediates the assembly of the influenza virus replicase.
Nature, 587, 2020
|
|
6PPN
 
 | | Structure of S. pombe Lsm2-8 with unprocessed U6 snRNA | | Descriptor: | Mimic of unprocessed U6 snRNA, Probable U6 snRNA-associated Sm-like protein LSm3, Probable U6 snRNA-associated Sm-like protein LSm4, ... | | Authors: | Montemayor, E.J, Butcher, S.E. | | Deposit date: | 2019-07-08 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Molecular basis for the distinct cellular functions of the Lsm1-7 and Lsm2-8 complexes.
Rna, 26, 2020
|
|
6YBJ
 
 | | Structure of MBP-Mcl-1 in complex with compound 3e | | Descriptor: | (2~{R})-2-[5-[3-chloranyl-2-methyl-4-[2-(4-methylpiperazin-1-yl)ethoxy]phenyl]-6-(5-fluoranylfuran-2-yl)thieno[2,3-d]pyrimidin-4-yl]oxy-3-[2-[(2-methylpyrazol-3-yl)methoxy]phenyl]propanoic acid, CHLORIDE ION, Maltose/maltodextrin-binding periplasmic protein,Induced myeloid leukemia cell differentiation protein Mcl-1, ... | | Authors: | Dokurno, P, Surgenor, A.E, Murray, J.B. | | Deposit date: | 2020-03-17 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of S64315, a Potent and Selective Mcl-1 Inhibitor.
J.Med.Chem., 63, 2020
|
|
6YD9
 
 | | Ecoli GyrB24 with inhibitor 16a | | Descriptor: | 1,2-ETHANEDIOL, DNA gyrase subunit B, N-[6-(3-azanylpropanoylamino)-1,3-benzothiazol-2-yl]-3,4-bis(chloranyl)-5-methyl-1H-pyrrole-2-carboxamide | | Authors: | Barancokova, M, Skok, Z, Benek, O, Cruz, C.D, Tammela, P, Tomasic, T, Zidar, N, Masic, L.P, Zega, A, Stevenson, C.E.M, Mundy, J, Lawson, D.M, Maxwell, A.M, Kikelj, D, Ilas, J. | | Deposit date: | 2020-03-20 | | Release date: | 2020-12-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Exploring the Chemical Space of Benzothiazole-Based DNA Gyrase B Inhibitors.
Acs Med.Chem.Lett., 11, 2020
|
|
6F3J
 
 | | The crystal structure of Glycogen Phosphorylase in complex with 10a | | Descriptor: | 4-[4-[5-[(2~{S},3~{R},4~{R},5~{S},6~{R})-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]-4~{H}-1,2,4-triazol-3-yl]phenyl]benzoic acid, Glycogen phosphorylase, muscle form, ... | | Authors: | Kyriakis, E, Stamati, E.C.V, Stravodimos, G.A, Skamnaki, V.T, Leonidas, D.D. | | Deposit date: | 2017-11-28 | | Release date: | 2018-02-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A multidisciplinary study of 3-( beta-d-glucopyranosyl)-5-substituted-1,2,4-triazole derivatives as glycogen phosphorylase inhibitors: Computation, synthesis, crystallography and kinetics reveal new potent inhibitors.
Eur J Med Chem, 147, 2018
|
|
6F3U
 
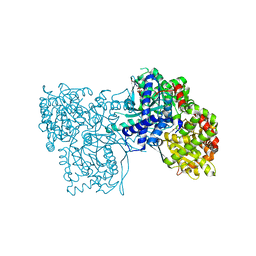 | | The crystal structure of Glycogen Phosphorylase in complex with 10h | | Descriptor: | (2~{R},3~{S},4~{R},5~{R},6~{S})-2-(hydroxymethyl)-6-(5-naphthalen-1-yl-4~{H}-1,2,4-triazol-3-yl)oxane-3,4,5-triol, Glycogen phosphorylase, muscle form, ... | | Authors: | Kyriakis, E, Gkerdi, A, Stravodimos, G.A, Skamnaki, V.T, Leonidas, D.D. | | Deposit date: | 2017-11-28 | | Release date: | 2018-02-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A multidisciplinary study of 3-( beta-d-glucopyranosyl)-5-substituted-1,2,4-triazole derivatives as glycogen phosphorylase inhibitors: Computation, synthesis, crystallography and kinetics reveal new potent inhibitors.
Eur J Med Chem, 147, 2018
|
|
6V3D
 
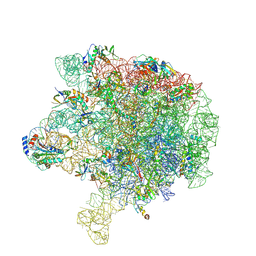 | |
7QNP
 
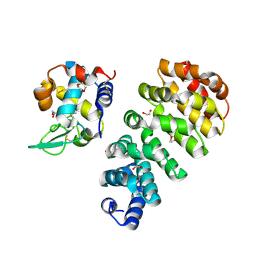 | | Designed Armadillo repeat protein N(A4)M4C(AII) co-crystallized with hen egg white lysozyme | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIETHYLENE DIOXIDE, Designed Armadillo Repeat Protein N(A4)M4C(AII), ... | | Authors: | Michel, E, Mittl, P.R.E, Plueckthun, A. | | Deposit date: | 2021-12-21 | | Release date: | 2022-06-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.586 Å) | | Cite: | Improved Repeat Protein Stability by Combined Consensus and Computational Protein Design.
Biochemistry, 62, 2023
|
|
6V6K
 
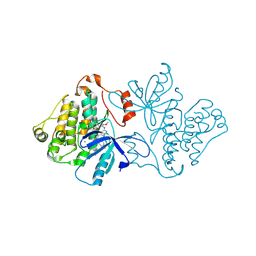 | | EGFR(T790M/V948R) in complex with LN2057 | | Descriptor: | CHLORIDE ION, Epidermal growth factor receptor, N-[3-({4-[4-(4-fluorophenyl)-2-(methylsulfanyl)-1H-imidazol-5-yl]pyridin-2-yl}amino)-4-methoxyphenyl]propanamide | | Authors: | Heppner, D.E, Eck, M.J. | | Deposit date: | 2019-12-05 | | Release date: | 2020-04-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for EGFR Mutant Inhibition by Trisubstituted Imidazole Inhibitors.
J.Med.Chem., 63, 2020
|
|
6P58
 
 | | Dark and Steady State-Illuminated Crystal Structure of Cyanobacteriochrome Receptor PixJ at 150K | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Methyl-accepting chemotaxis protein, ... | | Authors: | Clinger, J.A, Miller, M.D, Buirgie, E.S, Vierstra, R.D, Phillips Jr, G.N. | | Deposit date: | 2019-05-29 | | Release date: | 2019-12-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.499 Å) | | Cite: | Photoreversible interconversion of a phytochrome photosensory module in the crystalline state.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8GAL
 
 | | Crystal Structure of the E. coli LptA in complex with Murgantia histrionica Thanatin | | Descriptor: | Lipopolysaccharide export system protein LptA, Thanatin | | Authors: | Kibrom, A, Huynh, K, Donald, B, Zhou, P. | | Deposit date: | 2023-02-23 | | Release date: | 2023-07-19 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery, characterization, and redesign of potent antimicrobial thanatin orthologs from Chinavia ubica and Murgantia histrionica targeting E. coli LptA.
J Struct Biol X, 8, 2023
|
|
6YBG
 
 | | Structure of Mcl-1 in complex with compound 2g | | Descriptor: | (2~{R})-2-[5-[3-chloranyl-2-methyl-4-[2-(4-methylpiperazin-1-yl)ethoxy]phenyl]-6-(3-chlorophenyl)thieno[2,3-d]pyrimidin-4-yl]oxy-3-(2-methoxyphenyl)propanoic acid, CHLORIDE ION, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Dokurno, P, Surgenor, A.E, Murray, J.B. | | Deposit date: | 2020-03-17 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of S64315, a Potent and Selective Mcl-1 Inhibitor.
J.Med.Chem., 63, 2020
|
|
6N3S
 
 | | Crystal structure of apo-cruzain | | Descriptor: | 1,2-ETHANEDIOL, Cruzipain, PHOSPHATE ION | | Authors: | Silva, E.B, Dall, E, Rodrigues, F.T.G, Ferreira, R.S, Brandstetter, H. | | Deposit date: | 2018-11-16 | | Release date: | 2019-05-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.193 Å) | | Cite: | Cruzain structures: apocruzain and cruzain bound to S-methyl thiomethanesulfonate and implications for drug design.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
6F3R
 
 | | The crystal structure of Glycogen Phosphorylase in complex with 10c | | Descriptor: | (2~{S},3~{R},4~{R},5~{S},6~{R})-2-[5-(9~{H}-fluoren-2-yl)-4~{H}-1,2,4-triazol-3-yl]-6-(hydroxymethyl)oxane-3,4,5-triol, Glycogen phosphorylase, muscle form, ... | | Authors: | Kyriakis, E, Barkas, T.A, Stravodimos, G.A, Skamnaki, V.T, Leonidas, D.D. | | Deposit date: | 2017-11-28 | | Release date: | 2018-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A multidisciplinary study of 3-( beta-d-glucopyranosyl)-5-substituted-1,2,4-triazole derivatives as glycogen phosphorylase inhibitors: Computation, synthesis, crystallography and kinetics reveal new potent inhibitors.
Eur J Med Chem, 147, 2018
|
|
1FDP
 
 | | PROENZYME OF HUMAN COMPLEMENT FACTOR D, RECOMBINANT PROFACTOR D | | Descriptor: | PROENZYME OF COMPLEMENT FACTOR D | | Authors: | Jing, H, Macon, K.J, Moore, D, Delucas, L.J, Volanakis, J.E, Narayana, S.V.L. | | Deposit date: | 1998-12-03 | | Release date: | 1999-12-03 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of profactor D activation: from a highly flexible zymogen to a novel self-inhibited serine protease, complement factor D.
Embo J., 18, 1999
|
|
6F5B
 
 | |
6F5F
 
 | |
6F3S
 
 | | The crystal structure of Glycogen Phosphorylase in complex with 10d | | Descriptor: | (2~{R},3~{S},4~{R},5~{R},6~{S})-2-(hydroxymethyl)-6-[5-(4-phenylphenyl)-4~{H}-1,2,4-triazol-3-yl]oxane-3,4,5-triol, Glycogen phosphorylase, muscle form, ... | | Authors: | Kyriakis, E, Stamati, E.C.V, Stravodimos, G.A, Skamnaki, V.T, Leonidas, D.D. | | Deposit date: | 2017-11-28 | | Release date: | 2018-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A multidisciplinary study of 3-( beta-d-glucopyranosyl)-5-substituted-1,2,4-triazole derivatives as glycogen phosphorylase inhibitors: Computation, synthesis, crystallography and kinetics reveal new potent inhibitors.
Eur J Med Chem, 147, 2018
|
|
7TC3
 
 | | Human APE1 in the apo form | | Descriptor: | 1,2-ETHANEDIOL, DNA-(apurinic or apyrimidinic site) endonuclease, mitochondrial | | Authors: | Pidugu, L.S, Pozharski, E, Drohat, A.C. | | Deposit date: | 2021-12-22 | | Release date: | 2022-12-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.252 Å) | | Cite: | Characterizing inhibitors of human AP endonuclease 1.
Plos One, 18, 2023
|
|
7TC2
 
 | | Human APE1 in complex with 5-nitroindole-2-carboxylic acid | | Descriptor: | 1,2-ETHANEDIOL, 5-nitro-1H-indole-2-carboxylic acid, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Pidugu, L.S, Pozharski, E, Drohat, A.C. | | Deposit date: | 2021-12-22 | | Release date: | 2022-12-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Characterizing inhibitors of human AP endonuclease 1.
Plos One, 18, 2023
|
|
8G9Q
 
 | | Tricomplex of Compound-1, KRAS G12C, and CypA | | Descriptor: | GTPase KRas, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Tomlinson, A.C.A, Chen, A, Knox, J.E, Yano, J.K. | | Deposit date: | 2023-02-21 | | Release date: | 2023-08-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Chemical remodeling of a cellular chaperone to target the active state of mutant KRAS.
Science, 381, 2023
|
|
6OYU
 
 | | Structure of an ancestral-reconstructed cytochrome P450 1B1 with alpha-naphthoflavone | | Descriptor: | 2-PHENYL-4H-BENZO[H]CHROMEN-4-ONE, Cytochrome P450 1B1, GLYCEROL, ... | | Authors: | Bart, A.G, Harris, K.L, Scott, E.E. | | Deposit date: | 2019-05-15 | | Release date: | 2020-03-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structure of an ancestral mammalian family 1B1 cytochrome P450 with increased thermostability.
J.Biol.Chem., 295, 2020
|
|
8G9P
 
 | | Tricomplex of RMC-4998, KRAS G12C, and CypA | | Descriptor: | (2S)-2-{(5S)-7-[(2E)-4-(dimethylamino)-4-methylpent-2-enoyl]-1-oxo-2,7-diazaspiro[4.4]nonan-2-yl}-N-[(1P,8S,10R,14S,21M)-22-ethyl-21-{2-[(1S)-1-methoxyethyl]pyridin-3-yl}-18,18-dimethyl-9,15-dioxo-16-oxa-10,22,28-triazapentacyclo[18.5.2.1~2,6~.1~10,14~.0~23,27~]nonacosa-1(25),2(29),3,5,20,23,26-heptaen-8-yl]-3-methylbutanamide (non-preferred name), CHLORIDE ION, GTPase KRas, ... | | Authors: | Tomlinson, A.C.A, Saldajeno-Concar, M, Knox, J.E, Yano, J.K. | | Deposit date: | 2023-02-21 | | Release date: | 2023-08-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Chemical remodeling of a cellular chaperone to target the active state of mutant KRAS.
Science, 381, 2023
|
|
6P07
 
 | | Spastin hexamer in complex with substrate | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Sandate, C.R, Szyk, A, Zehr, E, Roll-Mecak, A, Lander, G.C. | | Deposit date: | 2019-05-16 | | Release date: | 2019-06-12 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | An allosteric network in spastin couples multiple activities required for microtubule severing.
Nat.Struct.Mol.Biol., 26, 2019
|
|
