4RAU
 
 | |
6A48
 
 | | Crystal structure of reelin N-terminal region | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Reelin | | Authors: | Nagae, M, Takagi, J. | | Deposit date: | 2018-06-19 | | Release date: | 2019-06-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural studies of reelin N-terminal region provides insights into a unique structural arrangement and functional multimerization.
J.Biochem., 169, 2021
|
|
4OJF
 
 | | Humanised 3D6 Fab complexed to amyloid beta 1-8 | | Descriptor: | Amyloid beta A4 protein, Humanised 3D6 Fab Heavy Chain, Humanised 3D6 Fab Light Chain | | Authors: | Miles, L.A, Crespi, G.A.N, Parker, M.W. | | Deposit date: | 2014-01-21 | | Release date: | 2015-01-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Crystallization and preliminary X-ray diffraction analysis of the Fab portion of the Alzheimer's disease immunotherapy candidate bapineuzumab complexed with amyloid-beta
ACTA CRYSTALLOGR.,SECT.F, 70, 2014
|
|
4OV5
 
 | | Structure of HLA-DR1 with a bound peptide with non-optimal alanine in the P1 pocket | | Descriptor: | HLA class I histocompatibility antigen, A-2 alpha chain, HLA class II histocompatibility antigen, ... | | Authors: | Trenh, P, Yin, L, Stern, L.J. | | Deposit date: | 2014-02-20 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.199 Å) | | Cite: | Susceptibility to HLA-DM Protein Is Determined by a Dynamic Conformation of Major Histocompatibility Complex Class II Molecule Bound with Peptide.
J.Biol.Chem., 289, 2014
|
|
1DN0
 
 | | STRUCTURE OF THE FAB FRAGMENT FROM A HUMAN IGM COLD AGGLUTININ | | Descriptor: | IGM-KAPPA COLD AGGLUTININ (HEAVY CHAIN), IGM-KAPPA COLD AGGLUTININ (LIGHT CHAIN) | | Authors: | Cauerhff, A, Braden, B, Carvalho, J.G, Leoni, J, Polikarpov, I, Goldbaum, F. | | Deposit date: | 1999-12-15 | | Release date: | 2001-01-24 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Three-dimensional structure of the Fab from a human IgM cold agglutinin.
J.Immunol., 165, 2000
|
|
4TSA
 
 | | Structure of a lysozyme FAb complex | | Descriptor: | FAb Heavy Chain, FAb Light Chain, Lysozyme C | | Authors: | Wensley, B. | | Deposit date: | 2014-06-18 | | Release date: | 2015-07-08 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structure of a lysozyme FAb complex
To Be Published
|
|
1H0T
 
 | | An affibody in complex with a target protein: structure and coupled folding | | Descriptor: | IMMUNOGLOBULIN G BINDING PROTEIN A, ZSPA-1 AFFIBODY | | Authors: | Wahlberg, E, Lendel, C, Helgstrand, M, Allard, P, Dincbas-Renqvist, V, Hedqvist, A, Berglund, H, Nygren, P.-A, Hard, T. | | Deposit date: | 2002-06-27 | | Release date: | 2003-02-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | An Affibody in Complex with a Target Protein: Structure and Coupled Folding
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1H2O
 
 | | SOLUTION STRUCTURE OF THE MAJOR CHERRY ALLERGEN PRU AV 1 MUTANT E45W | | Descriptor: | MAJOR ALLERGEN PRU AV 1 | | Authors: | Neudecker, P, Lehmann, K, Nerkamp, J, Schweimer, K, Sticht, H, Boehm, M, Scheurer, S, Vieths, S, Roesch, P. | | Deposit date: | 2002-08-12 | | Release date: | 2003-08-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Mutational Epitope Analysis of Pru Av 1 and Api G 1, the Major Allergens of Cherry (Prunus Avium) and Celery (Apium Graveolens): Correlating Ige Reactivity with Three-Dimensional Structure
Biochem.J., 376, 2003
|
|
1CG2
 
 | | CARBOXYPEPTIDASE G2 | | Descriptor: | CARBOXYPEPTIDASE G2, ZINC ION | | Authors: | Rowsell, S, Pauptit, R.A, Tucker, A.D, Melton, R.G, Blow, D.M, Brick, P. | | Deposit date: | 1996-12-20 | | Release date: | 1997-12-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of carboxypeptidase G2, a bacterial enzyme with applications in cancer therapy.
Structure, 5, 1997
|
|
7YD1
 
 | |
7A6O
 
 | |
7Y8J
 
 | | 3D1 in complex with 6-mer HR1 peptide from SARS-CoV-2 | | Descriptor: | Spike protein S2', heavy chain of 3D1, light chain of 3D1 | | Authors: | Lei, Y. | | Deposit date: | 2022-06-24 | | Release date: | 2023-07-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Cross-reactive epitopes between HIV and Coronavirus revealed by 3D1
To Be Published
|
|
6ZPL
 
 | | Inward-open structure of human glycine transporter 1 in complex with a benzoylisoindoline inhibitor, sybody Sb_GlyT1#7 and bound Na and Cl ions. | | Descriptor: | CHLORIDE ION, Endoglucanase H, SODIUM ION, ... | | Authors: | Shahsavar, A, Stohler, P, Bourenkov, G, Zimmermann, I, Siegrist, M, Guba, W, Pinard, E, Sinning, S, Seeger, M.A, Schneider, T.R, Dawson, R.J.P, Nissen, P. | | Deposit date: | 2020-07-08 | | Release date: | 2021-03-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.945 Å) | | Cite: | Structural insights into the inhibition of glycine reuptake.
Nature, 591, 2021
|
|
8GY5
 
 | |
7Z8O
 
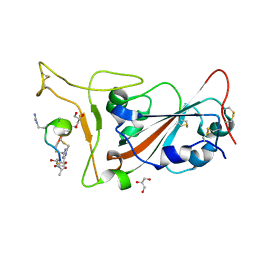 | | Crystal structure of SARS-CoV-2 S RBD in complex with a stapled peptide | | Descriptor: | 2,4,6-tris(chloromethyl)-1,3,5-triazine, GLYCEROL, Spike protein S1, ... | | Authors: | Brear, P, Chen, L, Gaynor, K, Harman, M, Dods, R, Hyvonen, M. | | Deposit date: | 2022-03-18 | | Release date: | 2023-06-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (0.96 Å) | | Cite: | Multivalent bicyclic peptides are an effective antiviral modality that can potently inhibit SARS-CoV-2.
Nat Commun, 14, 2023
|
|
6ZTD
 
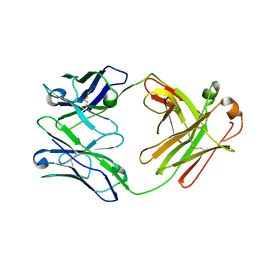 | | Crystal structure of the BCR Fab fragment from subset #169 case P6540 | | Descriptor: | Heavy chain of the Fab fragment from BCR derived from the P6540 CLL clone, Light chain of the Fab fragment from BCR derived from the P6540 CLL clone | | Authors: | Carriles, A.A, Minici, C, Degano, M. | | Deposit date: | 2020-07-18 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.43 Å) | | Cite: | Higher-order immunoglobulin repertoire restrictions in CLL: the illustrative case of stereotyped subsets 2 and 169.
Blood, 137, 2021
|
|
4ZNE
 
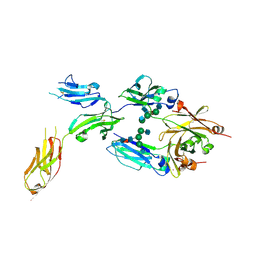 | | IgG1 Fc-FcgammaRI ecd complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Oganesyan, V.Y, Dall'Acqua, W.F. | | Deposit date: | 2015-05-04 | | Release date: | 2015-11-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structural insights into the interaction of human IgG1 with Fc gamma RI: no direct role of glycans in binding.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
8IA8
 
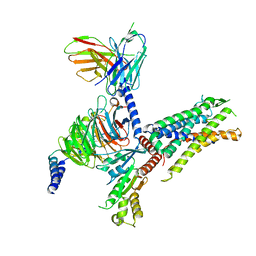 | | Cryo-EM structure of C3aR-Gi-scFv16 bound with E7 peptide | | Descriptor: | ALA-SER-LYS-LEU-GLY-LEU-ALA-ARG, C3a anaphylatoxin chemotactic receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Liao, Q, Chen, G, Du, Y, Ye, R.D. | | Deposit date: | 2023-02-08 | | Release date: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Cryo-EM structure of C3aR-Gi-scFv16 bound with E7 peptide
To Be Published
|
|
5ALC
 
 | | Ticagrelor antidote candidate Fab 72 in complex with ticagrelor | | Descriptor: | ANTI-TICAGRELOR FAB 72, HEAVY CHAIN, LIGHT CHAIN, ... | | Authors: | Buchanan, A, Newton, P, Pehrsson, S, Inghardt, T, Antonsson, T, Svensson, P, Sjogren, T, Oster, L, Janefeldt, A, Sandinge, A, Keyes, F, Austin, M, Spooner, J, Penney, M, Howells, G, Vaughan, T, Nylander, S. | | Deposit date: | 2015-03-07 | | Release date: | 2015-04-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and Functional Characterisation of a Specific Antidote for Ticagrelor.
Blood, 125, 2015
|
|
7YTE
 
 | |
7YTD
 
 | | Cryo-EM structure of four human FcmR bound to IgM-Fc/J | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fas apoptotic inhibitory molecule 3, Immunoglobulin J chain, ... | | Authors: | Li, Y, Shen, H, Xiao, J. | | Deposit date: | 2022-08-14 | | Release date: | 2023-02-01 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3.71 Å) | | Cite: | Immunoglobulin M perception by Fc mu R.
Nature, 615, 2023
|
|
7YSG
 
 | | Cryo-EM structure of human FcmR bound to sIgM | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fas apoptotic inhibitory molecule 3, ... | | Authors: | Li, Y, Shen, H, Xiao, J. | | Deposit date: | 2022-08-12 | | Release date: | 2023-02-01 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Immunoglobulin M perception by Fc mu R.
Nature, 615, 2023
|
|
7YTC
 
 | | Cryo-EM structure of human FcmR bound to IgM-Fc/J | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fas apoptotic inhibitory molecule 3, Immunoglobulin J chain, ... | | Authors: | Li, Y, Shen, H, Xiao, J. | | Deposit date: | 2022-08-14 | | Release date: | 2023-02-01 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Immunoglobulin M perception by Fc mu R.
Nature, 615, 2023
|
|
5B1C
 
 | | Crystal structure of DEN4 ED3 mutant with L387I | | Descriptor: | Envelope protein E, SULFATE ION | | Authors: | Kulkarni, M.R, Numoto, N, Ito, N, Kuroda, Y. | | Deposit date: | 2015-12-02 | | Release date: | 2016-02-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | Modeling and experimental assessment of a buried Leu-Ile mutation in dengue envelope domain III
Biochem.Biophys.Res.Commun., 471, 2016
|
|
7AQG
 
 | | Crystal Structure of Small Molecule Inhibitor TM5484 Bound to Stabilized Active Plasminogen Activator Inhibitor-1 (PAI-1-W175F) | | Descriptor: | 5-Chloro-2-[[2-[3-(furan-3-yl)anilino]-2-oxoacetyl]amino]benzoic acid, Plasminogen activator inhibitor 1, VHH-2g-42 (Nb42), ... | | Authors: | Sillen, M, Strelkov, S.V, Declerck, P.J. | | Deposit date: | 2020-10-21 | | Release date: | 2021-02-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structural Insight into the Two-Step Mechanism of PAI-1 Inhibition by Small Molecule TM5484.
Int J Mol Sci, 22, 2021
|
|
