3VCZ
 
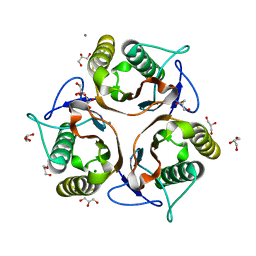 | | 1.80 Angstrom resolution crystal structure of a putative translation initiation inhibitor from Vibrio vulnificus CMCP6 | | Descriptor: | CALCIUM ION, Endoribonuclease L-PSP, GLYCEROL, ... | | Authors: | Halavaty, A.S, Minasov, G, Filippova, E.V, Dubrovska, I, Winsor, J, Shuvalova, L, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-01-04 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1.80 Angstrom resolution crystal structure of a putative translation initiation inhibitor from Vibrio vulnificus CMCP6
To be Published
|
|
4ZM9
 
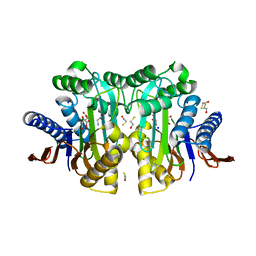 | | Crystal structure of circularly permuted human asparaginase-like protein 1 | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, BETA-MERCAPTOETHANOL, CITRATE ANION, ... | | Authors: | Li, W.Z, Zhang, Y. | | Deposit date: | 2015-05-02 | | Release date: | 2016-02-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Intramolecular Cleavage of the hASRGL1 Homodimer Occurs in Two Stages.
Biochemistry, 55, 2016
|
|
5XET
 
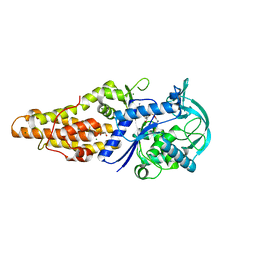 | | Crystal structure of Mycobacterium tuberculosis methionyl-tRNA synthetase bound by methionyl-adenylate (Met-AMP) | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Methionine--tRNA ligase, ... | | Authors: | Wang, W, Qin, B, Wojdyla, J.A, Wang, M, Gao, X, Cui, S. | | Deposit date: | 2017-04-06 | | Release date: | 2018-07-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structural characterization of free-state and product-stateMycobacterium tuberculosismethionyl-tRNA synthetase reveals an induced-fit ligand-recognition mechanism.
IUCrJ, 5, 2018
|
|
6QE8
 
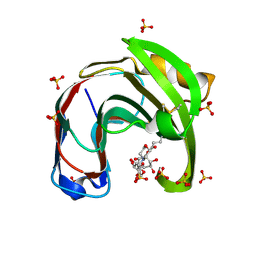 | | Crystal structure of Aspergillus niger GH11 endoxylanase XynA in complex with xylobiose epoxide activity based probe | | Descriptor: | (1~{R},3~{S},4~{R},5~{R})-5-[(2~{S},3~{R},4~{S},5~{R})-3,4,5-tris(oxidanyl)oxan-2-yl]oxycyclohexane-1,2,3,4-tetrol, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Endo-1,4-beta-xylanase A, ... | | Authors: | Wu, L, Rowland, R.J, Davies, G.J. | | Deposit date: | 2019-01-07 | | Release date: | 2019-06-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Dynamic and Functional Profiling of Xylan-Degrading Enzymes inAspergillusSecretomes Using Activity-Based Probes.
Acs Cent.Sci., 5, 2019
|
|
6UAR
 
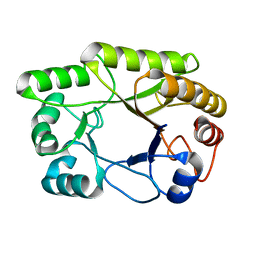 | | Crystal structure of a GH128 (subgroup I) endo-beta-1,3-glucanase from Amycolatopsis mediterranei (AmGH128_I) in complex with laminaritriose | | Descriptor: | Glyco_hydro_cc domain-containing protein, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose | | Authors: | Costa, P.A.C.R, Santos, C.R, Murakami, M.T. | | Deposit date: | 2019-09-11 | | Release date: | 2020-05-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural insights into beta-1,3-glucan cleavage by a glycoside hydrolase family.
Nat.Chem.Biol., 16, 2020
|
|
6B4O
 
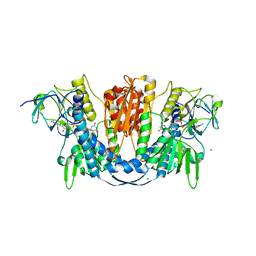 | | 1.73 Angstrom Resolution Crystal Structure of Glutathione Reductase from Enterococcus faecalis in Complex with FAD | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, Glutathione reductase, ... | | Authors: | Minasov, G, Warwzak, Z, Shuvalova, L, Dubrovska, I, Cardona-Correa, A, Grimshaw, S, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-09-27 | | Release date: | 2017-10-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | 1.73 Angstrom Resolution Crystal Structure of Glutathione Reductase from Enterococcus faecalis in Complex with FAD.
To Be Published
|
|
6F35
 
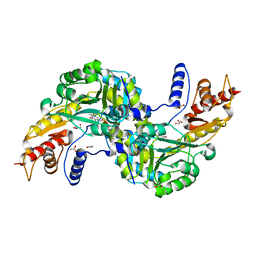 | | Crystal structure of the aspartate aminotranferase from Rhizobium meliloti | | Descriptor: | ACETATE ION, Aspartate aminotransferase B, GLYCEROL, ... | | Authors: | Cobessi, D, Graindorge, M, Giustini, C, Matringe, M. | | Deposit date: | 2017-11-28 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Tyrosine metabolism: identification of a key residue in the acquisition of prephenate aminotransferase activity by 1 beta aspartate aminotransferase.
Febs J., 286, 2019
|
|
6Q8N
 
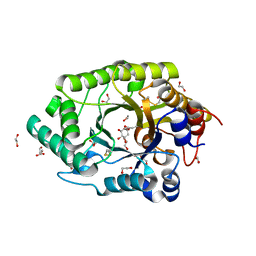 | | GH10 endo-xylanase in complex with xylobiose epoxide inhibitor | | Descriptor: | (1~{R},2~{S},4~{S},5~{R})-cyclohexane-1,2,3,4,5-pentol, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Davies, G.J, Rowland, R.J, Wu, L, Moroz, O, Blagova, E. | | Deposit date: | 2018-12-15 | | Release date: | 2019-06-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Dynamic and Functional Profiling of Xylan-Degrading Enzymes inAspergillusSecretomes Using Activity-Based Probes.
Acs Cent.Sci., 5, 2019
|
|
5KR8
 
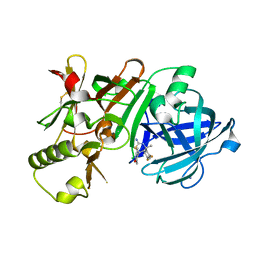 | | (4~{S},6~{S})-4-[2,4-bis(fluoranyl)phenyl]-6-(3,5-dimethyl-1,2-oxazol-4-yl)-4-methyl-5,6-dihydro-1,3-thiazin-2-amine (compound 5) bound to BACE1 | | Descriptor: | (4~{S},6~{S})-4-[2,4-bis(fluoranyl)phenyl]-6-(3,5-dimethyl-1,2-oxazol-4-yl)-4-methyl-5,6-dihydro-1,3-thiazin-2-amine, Beta-secretase 1, IODIDE ION | | Authors: | Lewis, H.A, Wu, Y.J, Rajamani, R, Thompson, L.A. | | Deposit date: | 2016-07-07 | | Release date: | 2016-09-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.118 Å) | | Cite: | Discovery of S3-Truncated, C-6 Heteroaryl Substituted Aminothiazine beta-Site APP Cleaving Enzyme-1 (BACE1) Inhibitors.
J.Med.Chem., 59, 2016
|
|
6H6Y
 
 | | GI.1 human norovirus protruding domain in complex with Nano-7 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Capsid protein VP1, ... | | Authors: | Kilic, T, Ruoff, K, Hansman, G.S. | | Deposit date: | 2018-07-30 | | Release date: | 2018-12-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structural Basis of Nanobodies Targeting the Prototype Norovirus.
J. Virol., 93, 2019
|
|
3I8A
 
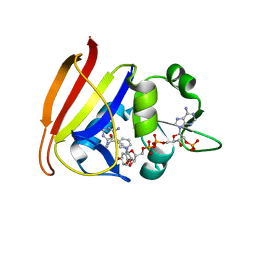 | | Staphylococcus aureus H30N, F98Y Dihydrofolate Reductase complexed with NADPH and 2,4-diamino-5-(3-(2,5-dimethoxyphenyl)prop-1-ynyl)-6-ethylpyrimidine (UCP120B) | | Descriptor: | 5-[3-(2,5-dimethoxyphenyl)prop-1-yn-1-yl]-6-ethylpyrimidine-2,4-diamine, Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Frey, K.M, Lombardo, M.N, Anderson, A.C. | | Deposit date: | 2009-07-09 | | Release date: | 2010-02-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Towards the understanding of resistance mechanisms in clinically isolated trimethoprim-resistant, methicillin-resistant Staphylococcus aureus dihydrofolate reductase.
J.Struct.Biol., 170, 2010
|
|
7F30
 
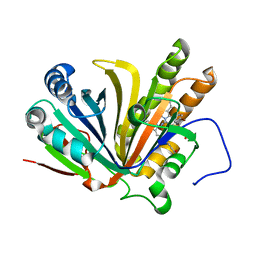 | | Crystal structure of OxdB E85A in complex with Z-2- (3-bromophenyl) propanal oxime | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Phenylacetaldoxime dehydratase, Z-2-(3-bromophenyl) propanal oxime | | Authors: | Muraki, N, Matsui, D, Asano, Y, Aono, S. | | Deposit date: | 2021-06-15 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structural analysis of aldoxime dehydratase from Bacillus sp. OxB-1: Importance of surface residues in optimization for crystallization.
J.Inorg.Biochem., 230, 2022
|
|
7F2Z
 
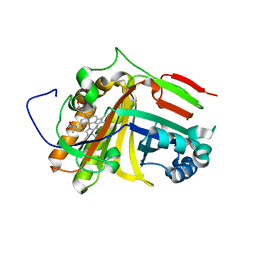 | | Crystal structure of OxdB E85A mutant (form II) | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Phenylacetaldoxime dehydratase | | Authors: | Muraki, N, Matsui, D, Asano, Y, Aono, S. | | Deposit date: | 2021-06-15 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structural analysis of aldoxime dehydratase from Bacillus sp. OxB-1: Importance of surface residues in optimization for crystallization.
J.Inorg.Biochem., 230, 2022
|
|
5NZQ
 
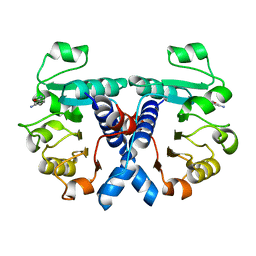 | | Crystal structure of human 3-phosphoglycerate dehydrogenase in complex with 3-(1,3-oxazol-5-yl)aniline. | | Descriptor: | 3-(1,3-oxazol-5-yl)aniline, D-3-phosphoglycerate dehydrogenase | | Authors: | Unterlass, J.E, Basle, A, Blackburn, T.J, Tucker, J, Cano, C, Noble, M.E.M, Curtin, N.J. | | Deposit date: | 2017-05-14 | | Release date: | 2017-06-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Validating and enabling phosphoglycerate dehydrogenase (PHGDH) as a target for fragment-based drug discovery in PHGDH-amplified breast cancer.
Oncotarget, 9, 2018
|
|
6FFM
 
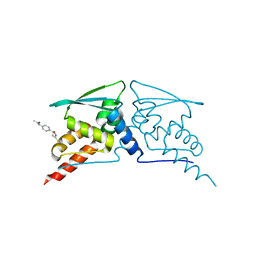 | |
7YPP
 
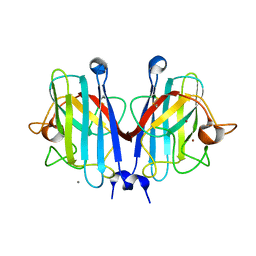 | | Structural basis of a superoxide dismutase from a tardigrade, Ramazzottius varieornatus strain YOKOZUNA-1. | | Descriptor: | CALCIUM ION, COPPER (II) ION, Superoxide dismutase [Cu-Zn], ... | | Authors: | Sim, K.-S, Fukuda, Y, Inoue, T. | | Deposit date: | 2022-08-04 | | Release date: | 2023-07-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of a superoxide dismutase from a tardigrade: Ramazzottius varieornatus strain YOKOZUNA-1.
Acta Crystallogr.,Sect.F, 79, 2023
|
|
5JCB
 
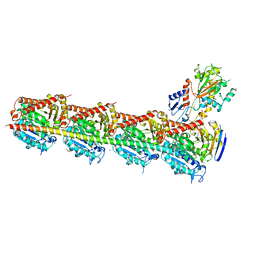 | | Microtubule depolymerizing agent podophyllotoxin derivative YJTSF1 | | Descriptor: | (5R,5aR,8aS,9R)-9-[(4H-1,2,4-triazol-3-yl)sulfanyl]-5-(3,4,5-trimethoxyphenyl)-5,8,8a,9-tetrahydro-2H-furo[3',4':6,7]naphtho[2,3-d][1,3]dioxol-6(5aH)-one, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Guan, Z, Zhao, W, Yin, P. | | Deposit date: | 2016-04-14 | | Release date: | 2017-09-27 | | Last modified: | 2025-09-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Insights into the Inhibition of Tubulin by the Antitumor Agent 4 beta-(1,2,4-triazol-3-ylthio)-4-deoxypodophyllotoxin.
ACS Chem. Biol., 12, 2017
|
|
3IS4
 
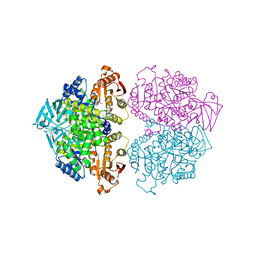 | | Crystal structure of Leishmania mexicana pyruvate kinase (LmPYK)in complex with 1,3,6,8-pyrenetetrasulfonic acid | | Descriptor: | GLYCEROL, Pyruvate kinase, pyrene-1,3,6,8-tetrasulfonic acid | | Authors: | Walkinshaw, M.D, Morgan, H.P. | | Deposit date: | 2009-08-25 | | Release date: | 2010-03-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | An improved strategy for the crystallization of Leishmania mexicana pyruvate kinase
Acta Crystallogr.,Sect.F, 66, 2010
|
|
5NUF
 
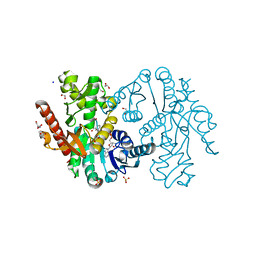 | | Cytosolic Malate Dehydrogenase 1 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Young, D, Messens, J, Huang, J, Reichheld, J.-P. | | Deposit date: | 2017-04-29 | | Release date: | 2018-01-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Self-protection of cytosolic malate dehydrogenase against oxidative stress in Arabidopsis.
J. Exp. Bot., 69, 2018
|
|
7F4C
 
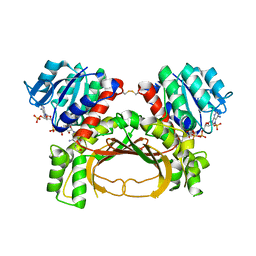 | | The crystal structure of the immature holo-enzyme of homoserine dehydrogenase complexed with NADP and 1,4-butandiol from the hyperthermophilic archaeon Sulfurisphaera tokodaii. | | Descriptor: | 1,4-BUTANEDIOL, Homoserine dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Ogata, K, Kaneko, R, Kubota, T, Watanabe, K, Kurihara, E, Oshima, T, Yoshimune, K, Goto, M. | | Deposit date: | 2021-06-18 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Conformational changes in the catalytic region are responsible for heat-induced activation of hyperthermophilic homoserine dehydrogenase.
Commun Biol, 5, 2022
|
|
6CGC
 
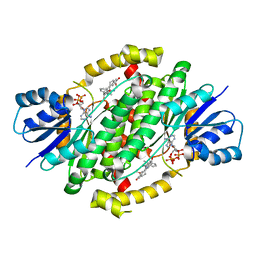 | | Crystal structure of human 17beta-HSD type 1 in ternary complex with 2-MeO-CC-156 and NADP+ | | Descriptor: | 3-{[(9beta,14beta,16alpha,17alpha)-3,17-dihydroxy-2-methoxyestra-1,3,5(10)-trien-16-yl]methyl}benzamide, Estradiol 17-beta-dehydrogenase 1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Li, T, Lin, S.X. | | Deposit date: | 2018-02-19 | | Release date: | 2019-01-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Combined Biophysical Chemistry Reveals a New Covalent Inhibitor with a Low-Reactivity Alkyl Halide.
J Phys Chem Lett, 9, 2018
|
|
5NZP
 
 | | Crystal structure of human 3-phosphoglycerate dehydrogenase in complex with 3-Hydroxybenzisoxazole | | Descriptor: | 1,2-benzoxazol-3-ol, D-3-phosphoglycerate dehydrogenase | | Authors: | Unterlass, J.E, Basle, A, Blackburn, T.J, Tucker, J, Cano, C, Noble, M.E.M, Curtin, N.J. | | Deposit date: | 2017-05-14 | | Release date: | 2017-06-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Validating and enabling phosphoglycerate dehydrogenase (PHGDH) as a target for fragment-based drug discovery in PHGDH-amplified breast cancer.
Oncotarget, 9, 2018
|
|
6FOD
 
 | | Vitamin D nuclear receptor complex 1 | | Descriptor: | (1~{R},3~{S},5~{Z})-4-methylidene-5-[(~{E})-3-[3-(6-methyl-6-oxidanyl-heptyl)phenyl]pent-2-enylidene]cyclohexane-1,3-diol, Nuclear receptor coactivator 1, Vitamin D3 receptor A | | Authors: | Rochel, N. | | Deposit date: | 2018-02-07 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Aromatic-Based Design of Highly Active and Noncalcemic Vitamin D Receptor Agonists.
J. Med. Chem., 61, 2018
|
|
6V1M
 
 | | Structure of NDM-1 bound to QPX7728 at 1.05 A | | Descriptor: | (1~{a}~{R},7~{b}~{S})-5-fluoranyl-2,2-bis(oxidanyl)-1~{a},7~{b}-dihydro-1~{H}-cyclopropa[c][1,2]benzoxaborinine-4-carboxylic acid, Metallo-beta-lactamase type 2, ZINC ION | | Authors: | Pemberton, O.A, Chen, Y. | | Deposit date: | 2019-11-20 | | Release date: | 2020-03-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Discovery of Cyclic Boronic Acid QPX7728, an Ultrabroad-Spectrum Inhibitor of Serine and Metallo-beta-lactamases.
J.Med.Chem., 63, 2020
|
|
5T6L
 
 | | Crystal structure of 10E8 Fab in complex with the MPER epitope scaffold T117v2 | | Descriptor: | 10E8 EPITOPE SCAFFOLD T117V2, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Antibody 10E8 FAB HEAVY CHAIN, ... | | Authors: | Irimia, A, Wilson, I.A. | | Deposit date: | 2016-09-01 | | Release date: | 2017-03-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Lipid interactions and angle of approach to the HIV-1 viral membrane of broadly neutralizing antibody 10E8: Insights for vaccine and therapeutic design.
PLoS Pathog., 13, 2017
|
|
