7SJ3
 
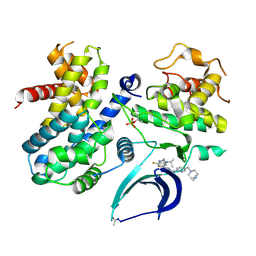 | | Structure of CDK4-Cyclin D3 bound to abemaciclib | | Descriptor: | Cyclin-dependent kinase 4, G1/S-specific cyclin-D3, N-{5-[(4-ethylpiperazin-1-yl)methyl]pyridin-2-yl}-5-fluoro-4-[4-fluoro-2-methyl-1-(propan-2-yl)-1H-benzimidazol-6-yl]py rimidin-2-amine | | Authors: | Hilgers, M.T, Pelletier, L.A. | | Deposit date: | 2021-10-15 | | Release date: | 2022-11-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structure of CDK4-Cyclin D3 bound to abemaciclib
To Be Published
|
|
7T1Y
 
 | | Structure of the Fbw7-Skp1-MycCdegron complex | | Descriptor: | F-box/WD repeat-containing protein 7, Myc proto-oncogene protein C terminal degron, S-phase kinase-associated protein 1, ... | | Authors: | Wang, B, Rusnac, D.V, Clurman, B.E, Zheng, N. | | Deposit date: | 2021-12-02 | | Release date: | 2022-02-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Two diphosphorylated degrons control c-Myc degradation by the Fbw7 tumor suppressor.
Sci Adv, 8, 2022
|
|
7TAD
 
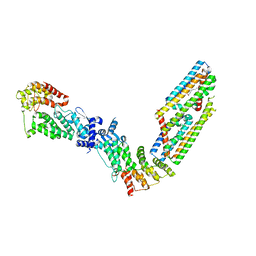 | | CryoEM structure of the (NPR1)2-(TGA3)2 complex | | Descriptor: | PALMITIC ACID, Regulatory protein NPR1, Transcription factor TGA3, ... | | Authors: | Wu, Q, Zhou, Y, Bartesaghi, A, Dong, X, Zhou, P. | | Deposit date: | 2021-12-20 | | Release date: | 2022-03-16 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis of NPR1 in activating plant immunity.
Nature, 605, 2022
|
|
7TAC
 
 | | Cryo-EM structure of the (TGA3)2-(NPR1)2-(TGA3)2 complex | | Descriptor: | PALMITIC ACID, Regulatory protein NPR1, Transcription factor TGA3, ... | | Authors: | Wu, Q, Zhou, Y, Bartesaghi, A, Dong, X, Zhou, P. | | Deposit date: | 2021-12-20 | | Release date: | 2022-03-16 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis of NPR1 in activating plant immunity.
Nature, 605, 2022
|
|
8W9F
 
 | |
8W9E
 
 | |
8W9D
 
 | |
6GU6
 
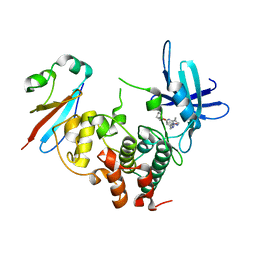 | | CDK1/Cks2 in complex with Dinaciclib | | Descriptor: | 3-[({3-ethyl-5-[(2S)-2-(2-hydroxyethyl)piperidin-1-yl]pyrazolo[1,5-a]pyrimidin-7-yl}amino)methyl]-1-hydroxypyridinium, Cyclin-dependent kinase 1, Cyclin-dependent kinases regulatory subunit 2 | | Authors: | Wood, D.J, Korolchuk, S, Tatum, N.J, Wang, L.Z, Endicott, J.A, Noble, M.E.M, Martin, M.P. | | Deposit date: | 2018-06-19 | | Release date: | 2018-12-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Differences in the Conformational Energy Landscape of CDK1 and CDK2 Suggest a Mechanism for Achieving Selective CDK Inhibition.
Cell Chem Biol, 26, 2019
|
|
6GU4
 
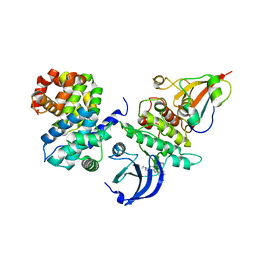 | | CDK1/CyclinB/Cks2 in complex with CGP74514A | | Descriptor: | Cyclin-dependent kinase 1, Cyclin-dependent kinases regulatory subunit 2, G2/mitotic-specific cyclin-B1, ... | | Authors: | Wood, D.J, Korolchuk, S, Tatum, N.J, Wang, L.Z, Endicott, J.A, Noble, M.E.M, Martin, M.P. | | Deposit date: | 2018-06-19 | | Release date: | 2018-12-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Differences in the Conformational Energy Landscape of CDK1 and CDK2 Suggest a Mechanism for Achieving Selective CDK Inhibition.
Cell Chem Biol, 26, 2019
|
|
6GU2
 
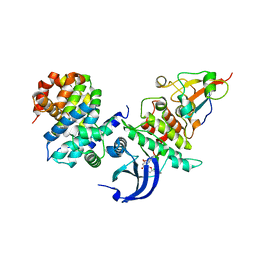 | | CDK1/CyclinB/Cks2 in complex with Flavopiridol | | Descriptor: | 2-(2-chlorophenyl)-8-[(3~{R},4~{R})-1-methyl-3-oxidanyl-piperidin-4-yl]-5,7-bis(oxidanyl)chromen-4-one, Cyclin-dependent kinase 1, Cyclin-dependent kinases regulatory subunit 2, ... | | Authors: | Wood, D.J, Korolchuk, S, Tatum, N.J, Wang, L.Z, Endicott, J.A, Noble, M.E.M, Martin, M.P. | | Deposit date: | 2018-06-19 | | Release date: | 2018-12-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Differences in the Conformational Energy Landscape of CDK1 and CDK2 Suggest a Mechanism for Achieving Selective CDK Inhibition.
Cell Chem Biol, 26, 2019
|
|
6GU3
 
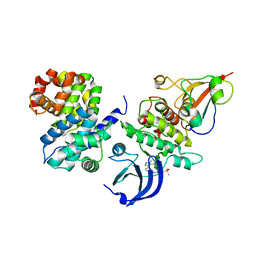 | | CDK1/CyclinB/Cks2 in complex with AZD5438 | | Descriptor: | 4-(2-methyl-3-propan-2-yl-imidazol-4-yl)-~{N}-(4-methylsulfonylphenyl)pyrimidin-2-amine, Cyclin-dependent kinase 1, Cyclin-dependent kinases regulatory subunit 2, ... | | Authors: | Wood, D.J, Korolchuk, S, Tatum, N.J, Wang, L.Z, Endicott, J.A, Noble, M.E.M, Martin, M.P. | | Deposit date: | 2018-06-19 | | Release date: | 2018-12-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Differences in the Conformational Energy Landscape of CDK1 and CDK2 Suggest a Mechanism for Achieving Selective CDK Inhibition.
Cell Chem Biol, 26, 2019
|
|
6R7N
 
 | | Structural basis of Cullin-2 RING E3 ligase regulation by the COP9 signalosome | | Descriptor: | COP9 signalosome complex subunit 1, COP9 signalosome complex subunit 2, COP9 signalosome complex subunit 3, ... | | Authors: | Faull, S.V, Lau, A.M.C, Martens, C, Ahdash, Z, Yebenes, H, Schmidt, C, Beuron, F, Cronin, N.B, Morris, E.P, Politis, A. | | Deposit date: | 2019-03-29 | | Release date: | 2019-08-28 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | Structural basis of Cullin 2 RING E3 ligase regulation by the COP9 signalosome.
Nat Commun, 10, 2019
|
|
6R7I
 
 | | Structural basis of Cullin-2 RING E3 ligase regulation by the COP9 signalosome | | Descriptor: | COP9 signalosome complex subunit 1, COP9 signalosome complex subunit 2, COP9 signalosome complex subunit 3, ... | | Authors: | Faull, S.F, Lau, A.M.C, Beuron, F, Cronin, N.B, Morris, E.P, Politis, A. | | Deposit date: | 2019-03-28 | | Release date: | 2019-08-28 | | Last modified: | 2019-09-04 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | Structural basis of Cullin 2 RING E3 ligase regulation by the COP9 signalosome.
Nat Commun, 10, 2019
|
|
6GU7
 
 | | CDK1/Cks2 in complex with AZD5438 | | Descriptor: | 4-(2-methyl-3-propan-2-yl-imidazol-4-yl)-~{N}-(4-methylsulfonylphenyl)pyrimidin-2-amine, Cyclin-dependent kinase 1, Cyclin-dependent kinases regulatory subunit 2 | | Authors: | Wood, D.J, Korolchuk, S, Tatum, N.J, Wang, L.Z, Endicott, J.A, Noble, M.E.M, Martin, M.P. | | Deposit date: | 2018-06-19 | | Release date: | 2018-12-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Differences in the Conformational Energy Landscape of CDK1 and CDK2 Suggest a Mechanism for Achieving Selective CDK Inhibition.
Cell Chem Biol, 26, 2019
|
|
8CG3
 
 | |
8CGH
 
 | |
6L1T
 
 | | Cryo-EM structure of phosphorylated Tyr39 a-synuclein amyloid fibril | | Descriptor: | Alpha-synuclein | | Authors: | Liu, C, Li, Y.M, Zhao, K, Lim, Y.J, Liu, Z.Y. | | Deposit date: | 2019-09-30 | | Release date: | 2020-08-12 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Parkinson's disease-related phosphorylation at Tyr39 rearranges alpha-synuclein amyloid fibril structure revealed by cryo-EM.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6L1U
 
 | | Cryo-EM structure of phosphorylated Tyr39 alpha-synuclein amyloid fibril | | Descriptor: | Alpha-synuclein | | Authors: | Liu, C, Li, Y.M, Zhao, K, Lim, Y.J, Liu, Z.Y. | | Deposit date: | 2019-09-30 | | Release date: | 2020-08-12 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Parkinson's disease-related phosphorylation at Tyr39 rearranges alpha-synuclein amyloid fibril structure revealed by cryo-EM.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6L0U
 
 | |
6SB2
 
 | | cryo-EM structure of mTORC1 bound to active RagA/C GTPases | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, Ras-related GTP-binding protein A, ... | | Authors: | Anandapadamanaban, M, Berndt, A, Masson, G.R, Perisic, O, Williams, R.L. | | Deposit date: | 2019-07-18 | | Release date: | 2019-10-16 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | Architecture of human Rag GTPase heterodimers and their complex with mTORC1.
Science, 366, 2019
|
|
6SB0
 
 | | cryo-EM structure of mTORC1 bound to PRAS40-fused active RagA/C GTPases | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, Proline-rich AKT1 substrate 1, ... | | Authors: | Anandapadamanaban, M, Berndt, A, Masson, G.R, Perisic, O, Williams, R.L. | | Deposit date: | 2019-07-18 | | Release date: | 2019-10-16 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | Architecture of human Rag GTPase heterodimers and their complex with mTORC1.
Science, 366, 2019
|
|
8HXX
 
 | | Cryo-EM structure of the histone deacetylase complex Rpd3S | | Descriptor: | Chromatin modification-related protein EAF3, Histone H3, Histone deacetylase RPD3, ... | | Authors: | Cui, H, Wang, H. | | Deposit date: | 2023-01-05 | | Release date: | 2023-09-27 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of histone deacetylase complex Rpd3S bound to nucleosome.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7SD0
 
 | | Cryo-EM structure of the SHOC2:PP1C:MRAS complex | | Descriptor: | Leucine-rich repeat protein SHOC-2, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Liau, N.P.D, Johnson, M.C, Hymowitz, S.G, Sudhamsu, J. | | Deposit date: | 2021-09-29 | | Release date: | 2022-04-20 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Structural basis for SHOC2 modulation of RAS signalling.
Nature, 609, 2022
|
|
7DTO
 
 | |
7E9F
 
 | | Cryo-EM structure of the 2:1 Orc1 BAH domain in complex with nucleosome | | Descriptor: | DNA (147-mer), Histone H2A.2, Histone H2B.2, ... | | Authors: | Jiang, H, Yu, C, Liu, C.P, Han, X, Yu, Z, Xu, R.M. | | Deposit date: | 2021-03-04 | | Release date: | 2022-09-07 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Nucleosome binding relinquishes the association of the BAH domain of Orc1 with Sir1
To Be Published
|
|
