6OXT
 
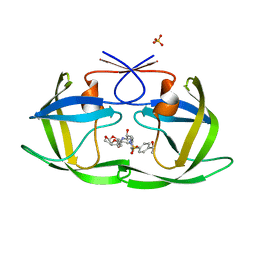 | | HIV-1 Protease NL4-3 WT in Complex with LR-84 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(2S,3R)-3-hydroxy-4-{({4-[(1S)-1-hydroxyethyl]phenyl}sulfonyl)[(2S)-2-methylbutyl]amino}-1-phenylbutan-2-yl]carbamate, Protease NL4-3, SULFATE ION | | Authors: | Lockbaum, G.J, Rusere, L.N, Lee, S.K, Henes, M, Kosovrasti, K, Spielvogel, E, Nalivaika, E.A, Swanstrom, R, KurtYilmaz, N, Schiffer, C.A, Ali, A. | | Deposit date: | 2019-05-14 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.861 Å) | | Cite: | HIV-1 Protease Inhibitors Incorporating Stereochemically Defined P2' Ligands To Optimize Hydrogen Bonding in the Substrate Envelope.
J.Med.Chem., 62, 2019
|
|
6OY0
 
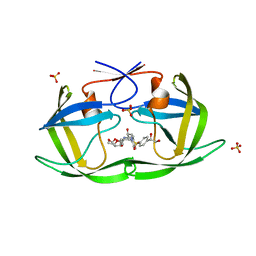 | | HIV-1 Protease NL4-3 WT in Complex with LR2-21 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(2S,3R)-4-{({4-[(1S)-1,2-dihydroxyethyl]phenyl}sulfonyl)[(2S)-2-methylbutyl]amino}-3-hydroxy-1-phenylbutan-2-yl]carbamate, Protease NL4-3, SULFATE ION | | Authors: | Lockbaum, G.J, Rusere, L.N, Lee, S.K, Henes, M, Kosovrasti, K, Spielvogel, E, Nalivaika, E.A, Swanstrom, R, KurtYilmaz, N, Schiffer, C.A, Ali, A. | | Deposit date: | 2019-05-14 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | HIV-1 Protease Inhibitors Incorporating Stereochemically Defined P2' Ligands To Optimize Hydrogen Bonding in the Substrate Envelope.
J.Med.Chem., 62, 2019
|
|
2EG8
 
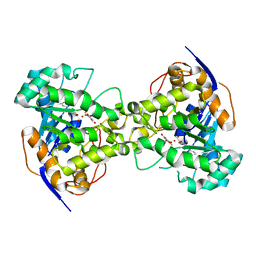 | | The crystal structure of E. coli dihydroorotase complexed with 5-fluoroorotic acid | | Descriptor: | 5-FLUORO-2,6-DIOXO-1,2,3,6-TETRAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, Dihydroorotase, ZINC ION | | Authors: | Lee, M, Maher, M.J, Guss, J.M. | | Deposit date: | 2007-02-28 | | Release date: | 2007-07-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of Ligand-free and Inhibitor Complexes of Dihydroorotase from Escherichia coli: Implications for Loop Movement in Inhibitor Design
J.Mol.Biol., 370, 2007
|
|
6ZLJ
 
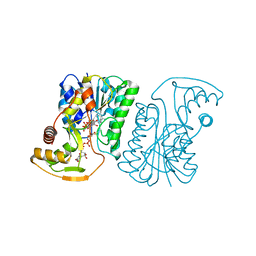 | | Crystal Structure of UDP-Glucuronic acid 4-epimerase Y149F mutant from Bacillus cereus in complex with UDP-4-DEOXY-4-FLUORO-Glucuronic acid and NAD | | Descriptor: | (2~{R},3~{S},4~{R},5~{R},6~{R})-6-[[[(2~{R},3~{S},4~{R},5~{R})-5-[2,4-bis(oxidanylidene)pyrimidin-1-yl]-3,4-bis(oxidany l)oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxy-3-fluoranyl-4,5-bis(oxidanyl)oxane-2-carboxylic acid, Epimerase domain-containing protein, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Iacovino, L.G, Mattevi, A. | | Deposit date: | 2020-06-30 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystallographic snapshots of UDP-glucuronic acid 4-epimerase ligand binding, rotation, and reduction.
J.Biol.Chem., 295, 2020
|
|
5ORB
 
 | | Crystal Structure of BAZ2B bromodomain in complex with 1-methyl-cyclopentapyrazole compound 30 | | Descriptor: | 1,2-ETHANEDIOL, 2-(4-methoxyphenyl)sulfanyl-~{N}-(2-methyl-5,6-dihydro-4~{H}-cyclopenta[c]pyrazol-3-yl)ethanamide, Bromodomain adjacent to zinc finger domain protein 2B | | Authors: | Lolli, G, Dalle Vedove, A, Marchand, J.-R, Caflisch, A. | | Deposit date: | 2017-08-15 | | Release date: | 2017-09-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.103 Å) | | Cite: | Discovery of Inhibitors of Four Bromodomains by Fragment-Anchored Ligand Docking.
J Chem Inf Model, 57, 2017
|
|
6H7Z
 
 | | X-ray structure of human glutamate carboxypeptidase II (GCPII) in complex with a inhibitor RNA 2-65-1 | | Descriptor: | (2~{S})-2-[[(2~{S})-6-[(6-fluoranylpyridin-3-yl)-methyl-amino]-1-oxidanyl-1,6-bis(oxidanylidene)hexan-2-yl]carbamoylamino]pentanedioic acid, 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Motlova, L, Novakova, Z, Barinka, C. | | Deposit date: | 2018-07-31 | | Release date: | 2018-12-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 2-Aminoadipic Acid-C(O)-Glutamate Based Prostate-Specific Membrane Antigen Ligands for Potential Use as Theranostics.
ACS Med Chem Lett, 9, 2018
|
|
3NYT
 
 | | X-ray crystal structure of the WlbE (WpbE) aminotransferase from pseudomonas aeruginosa, mutation K185A, in complex with the PLP external aldimine adduct with UDP-3-amino-2-N-acetyl-glucuronic acid, at 1.3 angstrom resolution | | Descriptor: | (2S,3S,4R,5R,6R)-5-(acetylamino)-6-{[(R)-{[(S)-{[(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methoxy}(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]oxy}-3-hydroxy-4-{[(1E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}tetrahydro-2H-pyran-2-carboxylic acid (non-preferred name), Aminotransferase WbpE, SODIUM ION | | Authors: | Holden, H.M, Thoden, J.B. | | Deposit date: | 2010-07-15 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.301 Å) | | Cite: | Structural investigation on WlaRG from Campylobacter jejuni: A sugar aminotransferase.
Protein Sci., 26, 2017
|
|
3V7D
 
 | | Crystal Structure of ScSkp1-ScCdc4-pSic1 peptide complex | | Descriptor: | Cell division control protein 4, Protein SIC1, Suppressor of kinetochore protein 1 | | Authors: | Tang, X, Orlicky, S, Mittag, T, Csizmok, V, Pawson, T, Forman-Kay, J, Sicheri, F, Tyers, M. | | Deposit date: | 2011-12-20 | | Release date: | 2012-05-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.306 Å) | | Cite: | Composite low affinity interactions dictate recognition of the cyclin-dependent kinase inhibitor Sic1 by the SCFCdc4 ubiquitin ligase.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3EUM
 
 | | A bimolecular anti-parallel-stranded Oxytricha nova telomeric quadruplex in complex with a 3,6-disubstituted acridine BSU-6066 | | Descriptor: | 3,6-Bis[3-(azepan-1-yl)propionamido]acridine, 5'-D(*DGP*DGP*DGP*DGP*DTP*DTP*DTP*DTP*DGP*DGP*DGP*DG)-3', POTASSIUM ION | | Authors: | Campbell, N.H, Parkinson, G, Neidle, S. | | Deposit date: | 2008-10-10 | | Release date: | 2008-10-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Selectivity in Ligand Recognition of G-Quadruplex Loops.
Biochemistry, 48, 2009
|
|
6MWK
 
 | |
6MWI
 
 | |
5T2T
 
 | | Crystal structure of Lymphocytic choriomeningitis mammarenavirus endonuclease bound to compound L742001 | | Descriptor: | (2Z)-4-[1-benzyl-4-(4-chlorobenzyl)piperidin-4-yl]-2-hydroxy-4-oxobut-2-enoic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Saez-Ayala, M, Yekwa, E.L, Canard, B, Alvarez, K, Ferron, F. | | Deposit date: | 2016-08-24 | | Release date: | 2017-09-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.967 Å) | | Cite: | Crystal structures ofLymphocytic choriomeningitis virusendonuclease domain complexed with diketo-acid ligands.
IUCrJ, 5, 2018
|
|
5OR9
 
 | | Crystal Structure of BAZ2B bromodomain in complex with 1-methyl-cyclopentapyrazole compound 13 | | Descriptor: | (2~{S})-1-(4-fluorophenyl)sulfonyl-~{N}-(2-methyl-5,6-dihydro-4~{H}-cyclopenta[c]pyrazol-3-yl)pyrrolidine-2-carboxamide, 1,2-ETHANEDIOL, Bromodomain adjacent to zinc finger domain protein 2B | | Authors: | Lolli, G, Vedove, A.D, Marchand, J.-R, Caflisch, A. | | Deposit date: | 2017-08-15 | | Release date: | 2017-09-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of Inhibitors of Four Bromodomains by Fragment-Anchored Ligand Docking.
J Chem Inf Model, 57, 2017
|
|
6U3K
 
 | | The crystal structure of 4-(pyridin-2-yl)benzoate-bound CYP199A4 | | Descriptor: | 4-(pyridin-2-yl)benzoic acid, CHLORIDE ION, Cytochrome P450, ... | | Authors: | Podgorski, M.N, Bruning, J.B, Bell, S.G. | | Deposit date: | 2019-08-21 | | Release date: | 2020-02-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Biophysical Techniques for Distinguishing Ligand Binding Modes in Cytochrome P450 Monooxygenases.
Biochemistry, 59, 2020
|
|
5AWK
 
 | | Crystal structure of VDR-LBD/partial agonist complex: 22S-ethyl analogue | | Descriptor: | (1R,3R)-5-[(2E)-2-[(1R,3aS,7aR)-1-[(2R,3S)-3-ethyl-5-oxidanyl-pentan-2-yl]-7a-methyl-2,3,3a,5,6,7-hexahydro-1H-inden-4-ylidene]ethylidene]-2-methylidene-cyclohexane-1,3-diol, Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 receptor,Vitamin D3 receptor | | Authors: | Anami, Y, Itoh, T, Yamamoto, K. | | Deposit date: | 2015-07-04 | | Release date: | 2015-11-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Fine tuning of agonistic/antagonistic activity for vitamin D receptor by 22-alkyl chain length of ligands: 22S-Hexyl compound unexpectedly restored agonistic activity.
Bioorg.Med.Chem., 23, 2015
|
|
3O2U
 
 | | S. cerevisiae Ubc12 | | Descriptor: | GLYCEROL, NEDD8-conjugating enzyme UBC12 | | Authors: | Scott, D.C, Monda, J.K, Grace, C.R.R, Duda, D.M, Kriwacki, R.W, Kurz, T, Schulman, B.A. | | Deposit date: | 2010-07-22 | | Release date: | 2010-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | A dual E3 mechanism for Rub1 ligation to Cdc53.
Mol.Cell, 39, 2010
|
|
6UD6
 
 | | Spectroscopic and structural characterization of a genetically encoded direct sensor for protein-ligand interactions | | Descriptor: | CHLORIDE ION, GLYCEROL, Streptavidin | | Authors: | Mills, J.H, Gleason, P.R, Simmons, C.R, Henderson, J.N, Kartchner, B.K. | | Deposit date: | 2019-09-18 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.502 Å) | | Cite: | Structural Origins of Altered Spectroscopic Properties upon Ligand Binding in Proteins Containing a Fluorescent Noncanonical Amino Acid.
Biochemistry, 60, 2021
|
|
5UTB
 
 | | Met form of sperm whale myoglobin V68A/I107Y | | Descriptor: | GLYCEROL, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Wang, B, Thomas, L.M, Richter-Addo, G.B. | | Deposit date: | 2017-02-14 | | Release date: | 2018-02-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Nitrosyl Myoglobins and Their Nitrite Precursors: Crystal Structural and Quantum Mechanics and Molecular Mechanics Theoretical Investigations of Preferred Fe -NO Ligand Orientations in Myoglobin Distal Pockets.
Biochemistry, 57, 2018
|
|
6UDC
 
 | | Spectroscopic and structural characterization of a genetically encoded direct sensor for protein-ligand interactions | | Descriptor: | BIOTIN, Streptavidin | | Authors: | Mills, J.H, Gleason, P.R, Simmons, C.R, Henderson, J.N, Kartchner, B.K. | | Deposit date: | 2019-09-19 | | Release date: | 2020-09-23 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Origins of Altered Spectroscopic Properties upon Ligand Binding in Proteins Containing a Fluorescent Noncanonical Amino Acid.
Biochemistry, 60, 2021
|
|
3G83
 
 | |
3D5V
 
 | |
5VZO
 
 | | Sperm whale myoglobin H64A with nitric oxide | | Descriptor: | GLYCEROL, Myoglobin, NITRIC OXIDE, ... | | Authors: | Wang, B, Thomas, L.M, Richter-Addo, G.B. | | Deposit date: | 2017-05-29 | | Release date: | 2018-06-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Nitrosyl Myoglobins and Their Nitrite Precursors: Crystal Structural and Quantum Mechanics and Molecular Mechanics Theoretical Investigations of Preferred Fe -NO Ligand Orientations in Myoglobin Distal Pockets.
Biochemistry, 57, 2018
|
|
1PWB
 
 | | High resolution crystal structure of an active recombinant fragment of human lung surfactant protein D with maltose | | Descriptor: | CALCIUM ION, Pulmonary surfactant-associated protein D, alpha-D-glucopyranose, ... | | Authors: | Shrive, A.K, Tharia, H.A, Strong, P, Kishore, U, Burns, I, Rizkallah, P.J, Reid, K.B, Greenhough, T.J. | | Deposit date: | 2003-07-01 | | Release date: | 2003-08-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | High-resolution structural insights into ligand binding and immune cell recognition by human lung surfactant protein D
J.Mol.Biol., 331, 2003
|
|
1Q0Y
 
 | | Anti-Morphine Antibody 9B1 Complexed with Morphine | | Descriptor: | (7R,7AS,12BS)-3-METHYL-2,3,4,4A,7,7A-HEXAHYDRO-1H-4,12-METHANO[1]BENZOFURO[3,2-E]ISOQUINOLINE-7,9-DIOL, Fab 9B1, Heavy chain, ... | | Authors: | Pozharski, E, Wilson, M.A, Hewagama, A, Shanafelt, A.B, Petsko, G, Ringe, D. | | Deposit date: | 2003-07-17 | | Release date: | 2004-04-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Anchoring a cationic ligand: the structure of the Fab fragment of the anti-morphine antibody 9B1 and its complex with morphine
J.Mol.Biol., 337, 2004
|
|
5VZQ
 
 | | Sperm whale myoglobin V68A/I107Y with nitric oxide | | Descriptor: | Myoglobin, NITRIC OXIDE, NITRITE ION, ... | | Authors: | Wang, B, Thomas, L.M, Richter-Addo, G.B. | | Deposit date: | 2017-05-29 | | Release date: | 2018-06-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Nitrosyl Myoglobins and Their Nitrite Precursors: Crystal Structural and Quantum Mechanics and Molecular Mechanics Theoretical Investigations of Preferred Fe -NO Ligand Orientations in Myoglobin Distal Pockets.
Biochemistry, 57, 2018
|
|
