8CNJ
 
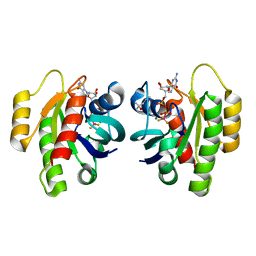 | | HRas(1-166) in complex with GDP and BeF3- | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, BERYLLIUM TRIFLUORIDE ION, GTPase HRas, ... | | Authors: | Baumann, P, Jin, Y. | | Deposit date: | 2023-02-23 | | Release date: | 2023-09-27 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Far-reaching effects of tyrosine64 phosphorylation on Ras revealed with BeF 3 - complexes.
Commun Chem, 7, 2024
|
|
8C0V
 
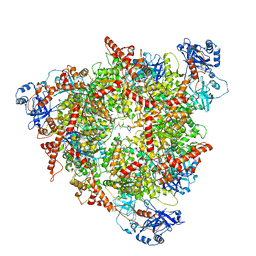 | | Structure of the peroxisomal Pex1/Pex6 ATPase complex bound to a substrate in single seam state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Ruettermann, M, Koci, M, Lill, P, Geladas, E.D, Kaschani, F, Klink, B.U, Erdmann, R, Gatsogiannis, C. | | Deposit date: | 2022-12-19 | | Release date: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structure of the peroxisomal Pex1/Pex6 ATPase complex bound to a substrate.
Nat Commun, 14, 2023
|
|
8C0W
 
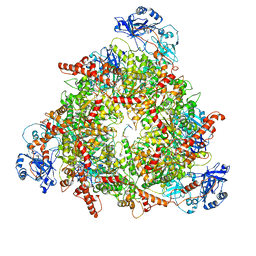 | | Structure of the peroxisomal Pex1/Pex6 ATPase complex bound to a substrate in twin seam state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Ruettermann, M, Koci, M, Lill, P, Geladas, E.D, Kaschani, F, Klink, B.U, Erdmann, R, Gatsogiannis, C. | | Deposit date: | 2022-12-19 | | Release date: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Structure of the peroxisomal Pex1/Pex6 ATPase complex bound to a substrate.
Nat Commun, 14, 2023
|
|
8CJ7
 
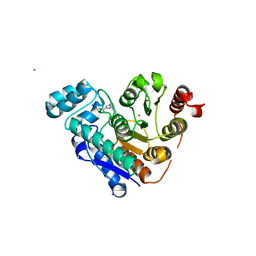 | | HDAC6 selective degraded (difluoromethyl)-1,3,4-oxadiazole substrate inhibitor | | Descriptor: | 6-[(5-pyridin-2-yl-1,2$l^{4},3,4-tetrazacyclopenta-1,3-dien-2-yl)methyl]pyridine-3-carbohydrazide, Histone deacetylase 6, IODIDE ION, ... | | Authors: | Sandmark, J, Ek, M, Ripa, L. | | Deposit date: | 2023-02-12 | | Release date: | 2023-10-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Selective and Bioavailable HDAC6 2-(Difluoromethyl)-1,3,4-oxadiazole Substrate Inhibitors and Modeling of Their Bioactivation Mechanism.
J.Med.Chem., 66, 2023
|
|
5GO9
 
 | |
5JN9
 
 | | Crystal structure for the complex of human carbonic anhydrase IV and ethoxyzolamide | | Descriptor: | 6-ethoxy-1,3-benzothiazole-2-sulfonamide, ACETATE ION, Carbonic anhydrase 4, ... | | Authors: | Chen, Z, Waheed, A, Di Cera, E, Sly, W.S. | | Deposit date: | 2016-04-29 | | Release date: | 2017-05-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Intrinsic thermodynamics of high affinity inhibitor binding to recombinant human carbonic anhydrase IV.
Eur. Biophys. J., 47, 2018
|
|
5JN8
 
 | | Crystal Structure for the complex of human carbonic anhydrase IV and acetazolamide | | Descriptor: | 5-ACETAMIDO-1,3,4-THIADIAZOLE-2-SULFONAMIDE, ACETATE ION, Carbonic anhydrase 4, ... | | Authors: | Chen, Z, Waheed, A, Di Cera, E, Sly, W.S. | | Deposit date: | 2016-04-29 | | Release date: | 2017-05-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Intrinsic thermodynamics of high affinity inhibitor binding to recombinant human carbonic anhydrase IV.
Eur. Biophys. J., 47, 2018
|
|
5FPN
 
 | | Structure of heat shock-related 70kDA protein 2 with small-molecule ligand 3,5-dimethyl-1H-pyrazole-4-carboxylic acid (AT9084) in an alternate binding site. | | Descriptor: | 3,5-DIMETHYL-1H-PYRAZOLE-4-CARBOXYLIC ACID, HEAT SHOCK-RELATED 70 KDA PROTEIN 2 | | Authors: | Jhoti, H, Ludlow, R.F, Patel, S, Saini, H.K, Tickle, I.J, Verdonk, M. | | Deposit date: | 2015-12-02 | | Release date: | 2015-12-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Detection of Secondary Binding Sites in Proteins Using Fragment Screening.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
1Z3J
 
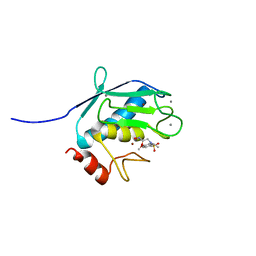 | | Solution Structure of MMP12 in the presence of N-isobutyl-N-4-methoxyphenylsulfonyl]glycyl hydroxamic acid (NNGH) | | Descriptor: | CALCIUM ION, Macrophage metalloelastase, N-ISOBUTYL-N-[4-METHOXYPHENYLSULFONYL]GLYCYL HYDROXAMIC ACID, ... | | Authors: | Bertini, I, Calderone, V, Cosenza, M, Fragai, M, Lee, Y.M, Luchinat, C, Mangani, S, Terni, B, Turano, P. | | Deposit date: | 2005-03-13 | | Release date: | 2005-04-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Conformational variability of matrix metalloproteinases: Beyond a single 3D structure.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
4XYD
 
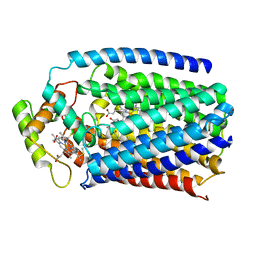 | | Nitric oxide reductase from Roseobacter denitrificans (RdNOR) | | Descriptor: | CALCIUM ION, COPPER (II) ION, FE (III) ION, ... | | Authors: | Crow, A. | | Deposit date: | 2015-02-02 | | Release date: | 2016-05-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure of the Membrane-intrinsic Nitric Oxide Reductase from Roseobacter denitrificans.
Biochemistry, 55, 2016
|
|
8W2O
 
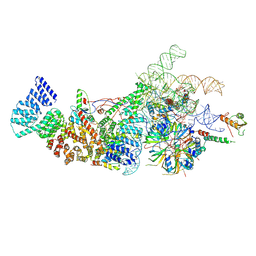 | | Yeast U1 snRNP with humanized U1C Zinc-Finger domain | | Descriptor: | 56 kDa U1 small nuclear ribonucleoprotein component, ACT1 pre-mRNA, Pre-mRNA-processing factor 39, ... | | Authors: | Shi, S.S, Kuang, Z.L, Zhao, R. | | Deposit date: | 2024-02-20 | | Release date: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Selected humanization of yeast U1 snRNP leads to global suppression of pre-mRNA splicing and mitochondrial dysfunction in the budding yeast.
Rna, 2024
|
|
2OSM
 
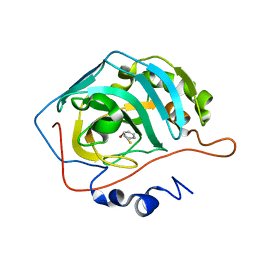 | | Inhibition of Carbonic Anhydrase II by Thioxolone: A Mechanistic and Structural Study | | Descriptor: | 2-MERCAPTOPHENOL, Carbonic anhydrase 2, ZINC ION | | Authors: | Albert, A.B, Caroli, G, Govindasamy, L, Agbandje-Mckenna, M, McKenna, R, Tripp, B.C. | | Deposit date: | 2007-02-06 | | Release date: | 2008-02-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Inhibition of Carbonic Anhydrase II by Thioxolone: A Mechanistic and Structural Study.
Biochemistry, 47, 2008
|
|
5FPR
 
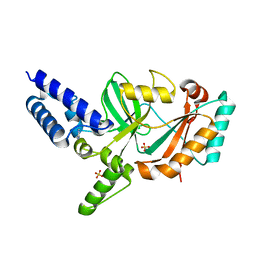 | | Structure of Bacterial DNA Ligase with small-molecule ligand pyrimidin-2-amine (AT371) in an alternate binding site. | | Descriptor: | DNA LIGASE, PYRIMIDIN-2-AMINE, SULFATE ION | | Authors: | Jhoti, H, Ludlow, R.F, Pathuri, P, Saini, H.K, Tickle, I.J, Tisi, D, Verdonk, M, Williams, P.A. | | Deposit date: | 2015-12-02 | | Release date: | 2015-12-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Detection of Secondary Binding Sites in Proteins Using Fragment Screening.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
1YR1
 
 | |
5GJW
 
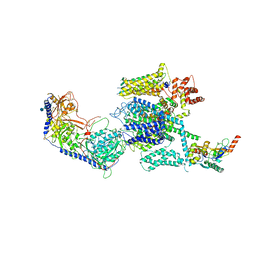 | | Structure of the mammalian voltage-gated calcium channel Cav1.1 complex for ClassII map | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wu, J.P, Yan, Z, Li, Z.Q, Zhou, Q, Yan, N. | | Deposit date: | 2016-07-02 | | Release date: | 2016-09-14 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure of the voltage-gated calcium channel Cav1.1 at 3.6 angstrom resolution
Nature, 537, 2016
|
|
8BQD
 
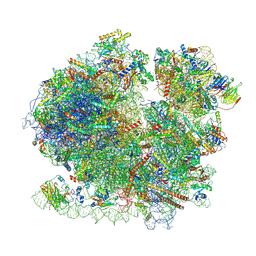 | | Yeast 80S ribosome in complex with Map1 (conformation 1) | | Descriptor: | 18S rRNA, 25S rRNA, 40S ribosomal protein S0-A, ... | | Authors: | Knorr, A.G, Mackens-Kiani, T, Musial, J, Berninghausen, O, Becker, T, Beatrix, B, Beckmann, R. | | Deposit date: | 2022-11-21 | | Release date: | 2023-03-22 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | The dynamic architecture of Map1- and NatB-ribosome complexes coordinates the sequential modifications of nascent polypeptide chains.
Plos Biol., 21, 2023
|
|
6TO9
 
 | |
4YCL
 
 | | Crystal structure of the SR CA2+-ATPASE with bound CPA | | Descriptor: | (6AR,11AS,11BR)-10-ACETYL-9-HYDROXY-7,7-DIMETHYL-2,6,6A,7,11A,11B-HEXAHYDRO-11H-PYRROLO[1',2':2,3]ISOINDOLO[4,5,6-CD]INDOL-11-ONE, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Ogawa, H, Takahashi, M, Kondou, Y, Toyoshima, C. | | Deposit date: | 2015-02-20 | | Release date: | 2015-04-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Interdomain communication in calcium pump as revealed in the crystal structures with transmembrane inhibitors
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
5JK4
 
 | | Phosphate-Binding Protein from Stenotrophomonas maltophilia. | | Descriptor: | Alkaline phosphatase, PHOSPHATE ION | | Authors: | Keegan, R, Waterman, D, Hopper, D, Coates, L, Guo, J, Coker, A.R, Erskine, P.T, Wood, S.P, Cooper, J.B. | | Deposit date: | 2016-04-25 | | Release date: | 2016-05-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | The 1.1 angstrom resolution structure of a periplasmic phosphate-binding protein from Stenotrophomonas maltophilia: a crystallization contaminant identified by molecular replacement using the entire Protein Data Bank.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
8TS0
 
 | |
8IQJ
 
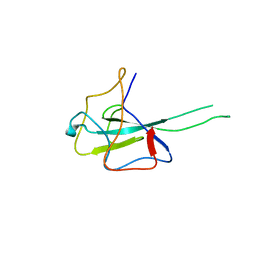 | | Crystal structure of SARS-CoV2 N-NTD | | Descriptor: | Nucleoprotein | | Authors: | Hong, J.Y, Hou, M.H. | | Deposit date: | 2023-03-16 | | Release date: | 2024-02-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Targeting protein-protein interaction interfaces with antiviral N protein inhibitor in SARS-CoV-2.
Biophys.J., 123, 2024
|
|
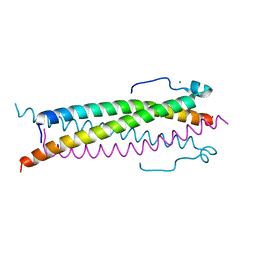
6TO5
 
 | |
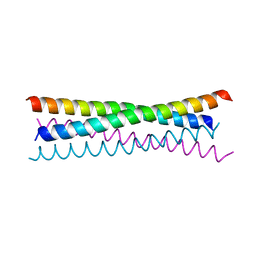
6TOC
 
 | |
6T7D
 
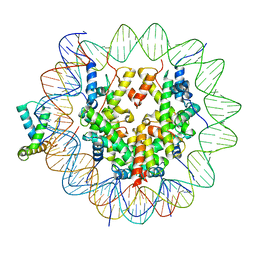 | | Structure of human Sox11 transcription factor in complex with a nucleosome | | Descriptor: | DNA (151-MER), Histone H2A type 1-B/E, Histone H2B type 1-K, ... | | Authors: | Dodonova, S.O, Zhu, F, Dienemann, C, Taipale, J, Cramer, P. | | Deposit date: | 2019-10-21 | | Release date: | 2020-04-29 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Nucleosome-bound SOX2 and SOX11 structures elucidate pioneer factor function.
Nature, 580, 2020
|
|
5E29
 
 | | Crystal Structure of Deoxygenated Hemoglobin in Complex with an Allosteric Effector and Nitric Oxide | | Descriptor: | 2-(nitrooxy)ethyl 2-(4-{2-[(3,5-dimethylphenyl)amino]-2-oxoethyl}phenoxy)-2-methylpropanoate, 2-{4-[(3,5-DIMETHYLANILINO)-CARBONYL-METHYL]-PHENOXY}-2-METHYLPROPIONIC ACID, CACODYLATE ION, ... | | Authors: | Safo, M.K, Deshpande, T.M. | | Deposit date: | 2015-09-30 | | Release date: | 2015-10-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Design, Synthesis, and Investigation of Novel Nitric Oxide (NO)-Releasing Prodrugs as Drug Candidates for the Treatment of Ischemic Disorders: Insights into NO-Releasing Prodrug Biotransformation and Hemoglobin-NO Biochemistry.
Biochemistry, 54, 2015
|
|
