5OVW
 
 | | Nanobody-bound BtuF, the vitamin B12 binding protein in Escherichia coli | | Descriptor: | GLYCEROL, Nanobody, Vitamin B12-binding protein | | Authors: | Mireku, S.A, Sauer, M.M, Glockshuber, R, Locher, K.P. | | Deposit date: | 2017-08-30 | | Release date: | 2017-11-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.653 Å) | | Cite: | Structural basis of nanobody-mediated blocking of BtuF, the cognate substrate-binding protein of the Escherichia coli vitamin B12 transporter BtuCD.
Sci Rep, 7, 2017
|
|
5SUT
 
 | | X-ray crystallographic structure of a covalent trimer derived from A-beta 17_36. Synchrotron data set. (ORN)CVFFCED(ORN)AII(SAR)L(ORN)V. | | Descriptor: | 16mer A-beta peptide: ORN-CYS-VAL-PHE-PHE-CYS-GLU-ASP-ORN-ALA-ILE-ILE-SAR-LEU-ORN-VAL, CHLORIDE ION | | Authors: | Kreutzer, A.G, Spencer, R.K, Nowick, J.S. | | Deposit date: | 2016-08-03 | | Release date: | 2017-01-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Stabilization, Assembly, and Toxicity of Trimers Derived from A beta.
J.Am.Chem.Soc., 139, 2017
|
|
5SUS
 
 | | X-ray crystallographic structure of a covalent trimer derived from A-beta 17_36. X-ray diffractometer data set. (ORN)CVF(MEA)CED(ORN)AIIGL(ORN)V. | | Descriptor: | 16mer A-beta peptide: ORN-CYS-VAL-PHE-MEA-CYS-GLU-ASP-ORN-ALA-ILE-ILE-GLY-LEU-ORN-VAL, CHLORIDE ION, SODIUM ION | | Authors: | Kreutzer, A.G, Yoo, S, Nowick, J.S. | | Deposit date: | 2016-08-03 | | Release date: | 2017-01-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.349 Å) | | Cite: | Stabilization, Assembly, and Toxicity of Trimers Derived from A beta.
J.Am.Chem.Soc., 139, 2017
|
|
5SUU
 
 | | X-ray crystallographic structure of a covalent trimer derived from A-beta 17-36. X-ray diffractometer data set. (ORN)CVFFCED(ORN)AII(SAR)L(ORN)V. | | Descriptor: | 16mer A-beta peptide: ORN-CYS-VAL-PHE-PHE-CYS-GLU-ASP-ORN-ALA-ILE-ILE-SAR-LEU-ORN-VAL, CHLORIDE ION, IODIDE ION | | Authors: | Kreutzer, A.G, Spencer, R.K, Nowick, J.S. | | Deposit date: | 2016-08-03 | | Release date: | 2017-01-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.032 Å) | | Cite: | Stabilization, Assembly, and Toxicity of Trimers Derived from A beta.
J.Am.Chem.Soc., 139, 2017
|
|
5SUR
 
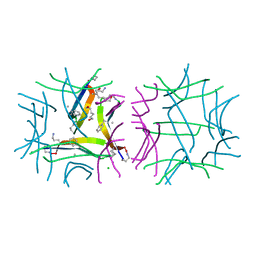 | | X-ray crystallographic structure of a covalent trimer derived from A-beta 17_36. Synchrotron data set. (ORN)CVF(MEA)CED(ORN)AIIGL(ORN)V. | | Descriptor: | 16mer A-beta peptide: ORN-CYS-VAL-PHE-MEA-CYS-GLU-ASP-ORN-ALA-ILE-ILE-GLY-LEU-ORN-VAL, CHLORIDE ION, HEXANE-1,6-DIOL, ... | | Authors: | Kreutzer, A.G, Yoo, S, Nowick, J.S. | | Deposit date: | 2016-08-03 | | Release date: | 2017-01-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Stabilization, Assembly, and Toxicity of Trimers Derived from A beta.
J.Am.Chem.Soc., 139, 2017
|
|
7Q1Z
 
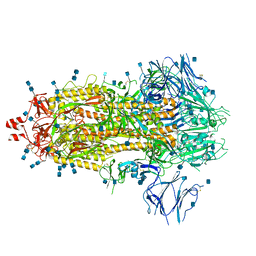 | | Structure of formaldehyde cross-linked SARS-CoV-2 S glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Sulbaran, G, Effantin, G, Schoehn, G, Weissenhorn, W. | | Deposit date: | 2021-10-22 | | Release date: | 2022-03-09 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Immunization with synthetic SARS-CoV-2 S glycoprotein virus-like particles protects macaques from infection.
Cell Rep Med, 3, 2022
|
|
7PGF
 
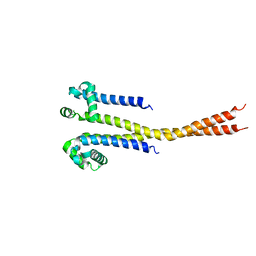 | |
7PGG
 
 | | NaVAb1p detergent (DM) | | Descriptor: | 2-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}ethyl heptadecanoate, Ion transport protein | | Authors: | Lolicato, M, Arrigoni, C. | | Deposit date: | 2021-08-13 | | Release date: | 2022-06-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Quaternary structure independent folding of voltage-gated ion channel pore domain subunits.
Nat.Struct.Mol.Biol., 29, 2022
|
|
2NCT
 
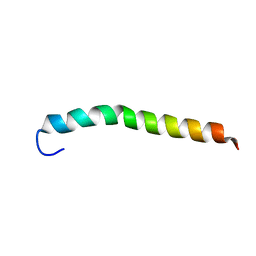 | | NMR assignment and structure of a peptide derived from the membrane proximal external region of HIV-1 gp41 in the presence of hexafluoroisopropanol | | Descriptor: | Envelope glycoprotein gp41 | | Authors: | Jimenez, M, Nieva, J.L, Rujas, E, Partida-Hanon, A, Bruix, M. | | Deposit date: | 2016-04-14 | | Release date: | 2017-02-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for broad neutralization of HIV-1 through the molecular recognition of 10E8 helical epitope at the membrane interface.
Sci Rep, 6, 2016
|
|
8PWH
 
 | | Atomic structure and conformational variability of the HER2-Trastuzumab-Pertuzumab complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Pertuzumab Fab heavy chain, Pertuzumab Fab light chain, ... | | Authors: | Ruedas, R, Vuillemot, R, Tubiana, T, Winter, J.M, Pieri, L, Arteni, A.A, Samson, C, Jonic, J, Mathieu, M, Bressanelli, S. | | Deposit date: | 2023-07-20 | | Release date: | 2024-02-21 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Structure and conformational variability of the HER2-trastuzumab-pertuzumab complex.
J.Struct.Biol., 216, 2024
|
|
2NCS
 
 | | NMR assignment and structure of a peptide derived from the membrane proximal external region of HIV-1 gp41 in the presence of dodecylphosphocholine micelles | | Descriptor: | Envelope glycoprotein gp41 | | Authors: | Jimenez, M, Nieva, J.L, Rujas, E, Partida-Hanon, A, Bruix, M. | | Deposit date: | 2016-04-14 | | Release date: | 2017-02-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for broad neutralization of HIV-1 through the molecular recognition of 10E8 helical epitope at the membrane interface.
Sci Rep, 6, 2016
|
|
5U5M
 
 | | CRYSTAL STRUCTURE OF I83E MEDITOPE-ENABLED TRASTUZUMAB WITH AZIDO-MEDITOPE | | Descriptor: | AZIDO-PEG4-MEDITOPE, Immunoglobulin G binding protein A, MEMAB TRASTUZUMAB, ... | | Authors: | Williams, J.C, Bzymek, K.P, Pucket, J, Avery, K.A, Ma, Y, Xie, J, Zer, C, Horne, D. | | Deposit date: | 2016-12-06 | | Release date: | 2018-03-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal Structure Of I83E Meditope-Enabled Trastuzumab With Azido-Meditope
To Be Published
|
|
8C6D
 
 | | Production of antigenically stable enterovirus A71 virus-like particles in Pichia pastoris as a vaccine candidate. | | Descriptor: | (2S,3R,4E)-2-aminooctadec-4-ene-1,3-diol, Genome polyprotein, Genome polyprotein (Fragment) | | Authors: | Kingston, N.J, Snowden, J.S, Stonehouse, N.J, Rowlands, D.J, Hogle, J.M. | | Deposit date: | 2023-01-11 | | Release date: | 2023-02-22 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Production of antigenically stable enterovirus A71 virus-like particles in Pichia pastoris as a vaccine candidate.
Biorxiv, 2023
|
|
7SSC
 
 | |
5U6A
 
 | | CRYSTAL STRUCTURE OF I83E MEDITOPE-ENABLED TRASTUZUMAB WITH AZIDO-PEG3-MEDITOPE | | Descriptor: | Heavy Chain, Immunoglobulin G binding protein A, Light Chain, ... | | Authors: | Williams, J.C, Bzymek, K.P, Pucket, J, Avery, K.A, Ma, Y, Xie, J, Zer, C, Horne, D. | | Deposit date: | 2016-12-07 | | Release date: | 2018-03-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.736 Å) | | Cite: | Crystal Structure Of I83E Meditope-Enabled Trastuzumab With Azido-PEG3-Meditope
To Be Published
|
|
3RRT
 
 | |
3RRR
 
 | | Structure of the RSV F protein in the post-fusion conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fusion glycoprotein F0 | | Authors: | McLellan, J.S, Yongping, Y, Graham, B.S, Kwong, P.D. | | Deposit date: | 2011-04-30 | | Release date: | 2011-06-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.821 Å) | | Cite: | Structure of respiratory syncytial virus fusion glycoprotein in the postfusion conformation reveals preservation of neutralizing epitopes.
J.Virol., 85, 2011
|
|
5VYH
 
 | | Crystal Structure of MERS-CoV S1 N-terminal Domain | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, FOLIC ACID, ... | | Authors: | Wang, N, Wrapp, D, Pallesen, J, Ward, A.B, McLellan, J.S. | | Deposit date: | 2017-05-25 | | Release date: | 2017-08-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Immunogenicity and structures of a rationally designed prefusion MERS-CoV spike antigen.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
4MA7
 
 | | Crystal structure of mouse prion protein complexed with Promazine | | Descriptor: | Major prion protein, POM1 heavy chain, POM1 light chain, ... | | Authors: | Baral, P.K, Swayampakula, M, James, M.N.G. | | Deposit date: | 2013-08-15 | | Release date: | 2014-01-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural basis of prion inhibition by phenothiazine compounds.
Structure, 22, 2014
|
|
4MA8
 
 | | Crystal structure of mouse prion protein complexed with Chlorpromazine | | Descriptor: | 3-(2-chloro-10H-phenothiazin-10-yl)-N,N-dimethylpropan-1-amine, Major prion protein, POM1 heavy chain, ... | | Authors: | Baral, P.K, Swayampakula, M, James, M.N.G. | | Deposit date: | 2013-08-15 | | Release date: | 2014-01-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of prion inhibition by phenothiazine compounds.
Structure, 22, 2014
|
|
4M61
 
 | |
8BPG
 
 | | FcMR binding at subunit Fcu3 of IgM pentamer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fas apoptotic inhibitory molecule 3, ... | | Authors: | Chen, Q, Rosenthal, P, Tolar, P. | | Deposit date: | 2022-11-16 | | Release date: | 2023-04-12 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for Fc receptor recognition of immunoglobulin M.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8BPF
 
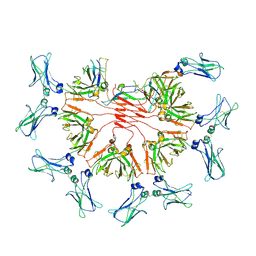 | | FcMR binding at subunit Fcu1 of IgM pentamer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fas apoptotic inhibitory molecule 3, ... | | Authors: | Chen, Q, Rosenthal, P, Tolar, P. | | Deposit date: | 2022-11-16 | | Release date: | 2023-04-12 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis for Fc receptor recognition of immunoglobulin M.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8BPE
 
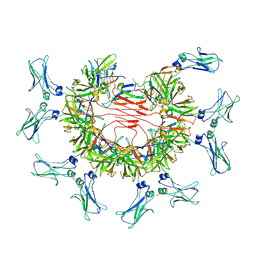 | | 8:1 binding of FcMR on IgM pentameric core | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fas apoptotic inhibitory molecule 3, ... | | Authors: | Chen, Q, Rosenthal, P, Tolar, P. | | Deposit date: | 2022-11-16 | | Release date: | 2023-04-12 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (3.63 Å) | | Cite: | Structural basis for Fc receptor recognition of immunoglobulin M.
Nat.Struct.Mol.Biol., 30, 2023
|
|
5N2F
 
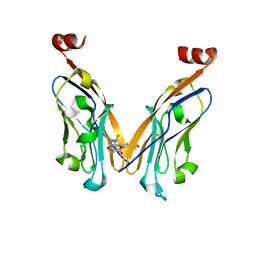 | | Structure of PD-L1/small-molecule inhibitor complex | | Descriptor: | 4-[[4-[[3-(2,3-dihydro-1,4-benzodioxin-6-yl)-2-methyl-phenyl]methoxy]-2,5-bis(fluoranyl)phenyl]methylamino]-3-oxidanylidene-butanoic acid, Programmed cell death 1 ligand 1 | | Authors: | Guzik, K, Zak, K.M, Grudnik, P, Dubin, G, Holak, T.A. | | Deposit date: | 2017-02-07 | | Release date: | 2017-06-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Small-Molecule Inhibitors of the Programmed Cell Death-1/Programmed Death-Ligand 1 (PD-1/PD-L1) Interaction via Transiently Induced Protein States and Dimerization of PD-L1.
J. Med. Chem., 60, 2017
|
|
