1FLC
 
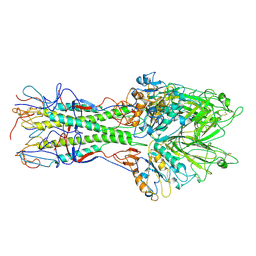 | | X-RAY STRUCTURE OF THE HAEMAGGLUTININ-ESTERASE-FUSION GLYCOPROTEIN OF INFLUENZA C VIRUS | | Descriptor: | HAEMAGGLUTININ-ESTERASE-FUSION GLYCOPROTEIN, alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Rosenthal, P.B, Zhang, X, Formanowski, F, Fitz, W, Wong, C.H, Meier-Ewert, H, Skehel, J.J, Wiley, D.C. | | Deposit date: | 1999-02-22 | | Release date: | 2000-03-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of the haemagglutinin-esterase-fusion glycoprotein of influenza C virus.
Nature, 396, 1998
|
|
8R5I
 
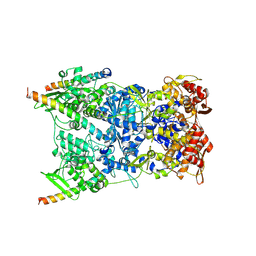 | | In situ structure of the Vaccinia virus (WR) A4/A10 palisade trimer in mature virions by flexible fitting into a cryoET map | | Descriptor: | Core protein A10, Core protein A4 | | Authors: | Calcraft, T, Hernandez-Gonzalez, M, Nans, A, Rosenthal, P.B, Way, M. | | Deposit date: | 2023-11-16 | | Release date: | 2024-01-10 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (9.7 Å) | | Cite: | Palisade structure in intact vaccinia virions.
Mbio, 15, 2024
|
|
8BPF
 
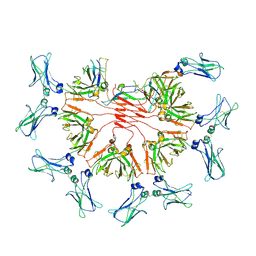 | | FcMR binding at subunit Fcu1 of IgM pentamer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fas apoptotic inhibitory molecule 3, ... | | Authors: | Chen, Q, Rosenthal, P, Tolar, P. | | Deposit date: | 2022-11-16 | | Release date: | 2023-04-12 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis for Fc receptor recognition of immunoglobulin M.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8BPE
 
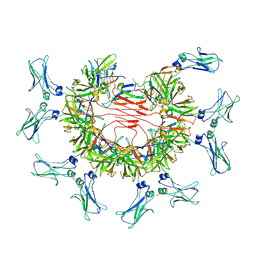 | | 8:1 binding of FcMR on IgM pentameric core | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fas apoptotic inhibitory molecule 3, ... | | Authors: | Chen, Q, Rosenthal, P, Tolar, P. | | Deposit date: | 2022-11-16 | | Release date: | 2023-04-12 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (3.63 Å) | | Cite: | Structural basis for Fc receptor recognition of immunoglobulin M.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8BPG
 
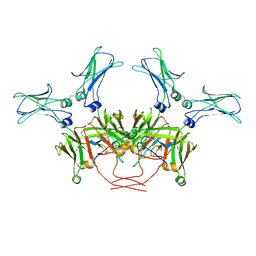 | | FcMR binding at subunit Fcu3 of IgM pentamer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fas apoptotic inhibitory molecule 3, ... | | Authors: | Chen, Q, Rosenthal, P, Tolar, P. | | Deposit date: | 2022-11-16 | | Release date: | 2023-04-12 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for Fc receptor recognition of immunoglobulin M.
Nat.Struct.Mol.Biol., 30, 2023
|
|
6N1Q
 
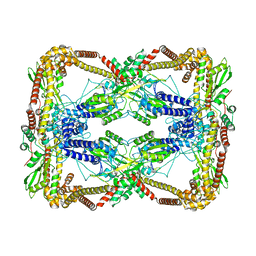 | | Dihedral oligomeric complex of GyrA N-terminal fragment, solved by cryoEM in D2 symmetry | | Descriptor: | DNA gyrase subunit A | | Authors: | Soczek, K.M, Grant, T, Rosenthal, P.B, Mondragon, A. | | Deposit date: | 2018-11-10 | | Release date: | 2018-12-05 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (5.16 Å) | | Cite: | CryoEM structures of open dimers of Gyrase A in complex with DNA illuminate mechanism of strand passage.
Elife, 7, 2018
|
|
7QDO
 
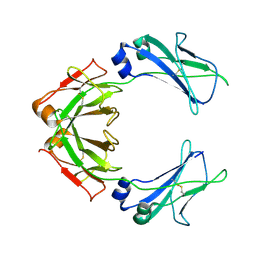 | | Cryo-EM structure of human monomeric IgM-Fc | | Descriptor: | Isoform 2 of Immunoglobulin heavy constant mu | | Authors: | Chen, Q, Rosenthal, P, Tolar, P. | | Deposit date: | 2021-11-27 | | Release date: | 2022-10-26 | | Last modified: | 2022-11-09 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryomicroscopy reveals the structural basis for a flexible hinge motion in the immunoglobulin M pentamer.
Nat Commun, 13, 2022
|
|
8AE2
 
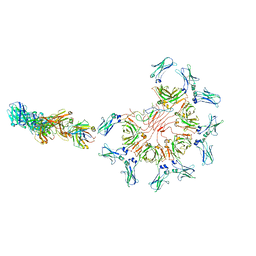 | | Cryo-EM structure of full-length human immunoglobulin M - F(ab')2 conformation 5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, IgM C2-domain from mouse, ... | | Authors: | Chen, Q, Rosenthal, P, Tolar, P. | | Deposit date: | 2022-07-12 | | Release date: | 2022-10-26 | | Last modified: | 2022-11-09 | | Method: | ELECTRON MICROSCOPY (8.5 Å) | | Cite: | Cryomicroscopy reveals the structural basis for a flexible hinge motion in the immunoglobulin M pentamer.
Nat Commun, 13, 2022
|
|
8ADZ
 
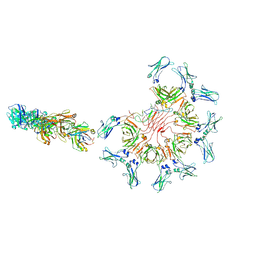 | | Cryo-EM structure of full-length human immunoglobulin M - F(ab')2 conformation 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, IgM C2-domain from mouse, ... | | Authors: | Chen, Q, Rosenthal, P, Tolar, P. | | Deposit date: | 2022-07-12 | | Release date: | 2022-10-26 | | Last modified: | 2022-11-09 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | Cryomicroscopy reveals the structural basis for a flexible hinge motion in the immunoglobulin M pentamer.
Nat Commun, 13, 2022
|
|
8AE0
 
 | | Cryo-EM structure of full-length human immunoglobulin M - F(ab')2 conformation 3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, IgM C2-domain from mouse, ... | | Authors: | Chen, Q, Rosenthal, P, Tolar, P. | | Deposit date: | 2022-07-12 | | Release date: | 2022-10-26 | | Last modified: | 2022-11-09 | | Method: | ELECTRON MICROSCOPY (7.1 Å) | | Cite: | Cryomicroscopy reveals the structural basis for a flexible hinge motion in the immunoglobulin M pentamer.
Nat Commun, 13, 2022
|
|
8AE3
 
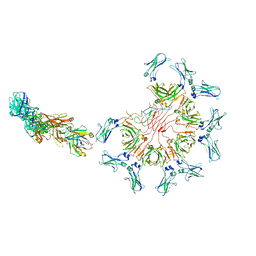 | | Cryo-EM structure of full-length human immunoglobulin M - F(ab')2 conformation 4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, IgM C2-domain from mouse, ... | | Authors: | Chen, Q, Rosenthal, P, Tolar, P. | | Deposit date: | 2022-07-12 | | Release date: | 2022-10-26 | | Last modified: | 2022-11-09 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | Cryomicroscopy reveals the structural basis for a flexible hinge motion in the immunoglobulin M pentamer.
Nat Commun, 13, 2022
|
|
8ADY
 
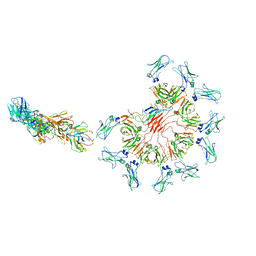 | | Cryo-EM structure of full-length human immunoglobulin M - F(ab')2 conformation 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, IgM C2-domain from mouse, ... | | Authors: | Chen, Q, Rosenthal, P, Tolar, P. | | Deposit date: | 2022-07-12 | | Release date: | 2022-10-26 | | Last modified: | 2022-11-09 | | Method: | ELECTRON MICROSCOPY (5.2 Å) | | Cite: | Cryomicroscopy reveals the structural basis for a flexible hinge motion in the immunoglobulin M pentamer.
Nat Commun, 13, 2022
|
|
6HJR
 
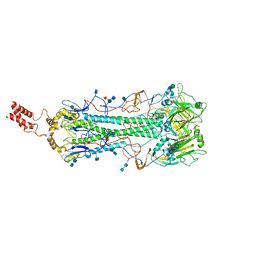 | |
6HJN
 
 | | Structure of Influenza Hemagglutinin ectodomain (A/duck/Alberta/35/76) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, ... | | Authors: | Benton, D.J, Rosenthal, P.B. | | Deposit date: | 2018-09-04 | | Release date: | 2018-09-26 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Influenza hemagglutinin membrane anchor.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
7AWA
 
 | | SctV (SsaV) cytoplasmic domain | | Descriptor: | Secretion system apparatus protein SsaV | | Authors: | Matthews-Palmer, T.R.S, Gonzalez-Rodriguez, N, Calcraft, T, Lagercrantz, S, Zachs, T, Yu, X.J, Grabe, G, Holden, D, Nans, A, Rosenthal, P, Rouse, S, Beeby, M. | | Deposit date: | 2020-11-06 | | Release date: | 2021-04-14 | | Last modified: | 2021-04-21 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of the cytoplasmic domain of SctV (SsaV) from the Salmonella SPI-2 injectisome and implications for a pH sensing mechanism.
J.Struct.Biol., 213, 2021
|
|
6HJQ
 
 | |
6HJP
 
 | |
7BNN
 
 | | Open conformation of D614G SARS-CoV-2 spike with 1 Erect RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Benton, D.J, Wrobel, A.G, Rosenthal, P.B, Gamblin, S.J. | | Deposit date: | 2021-01-22 | | Release date: | 2021-02-03 | | Last modified: | 2021-03-10 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The effect of the D614G substitution on the structure of the spike glycoprotein of SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7BBH
 
 | | Structure of Coronavirus Spike from Smuggled Guangdong Pangolin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Surface glycoprotein | | Authors: | Wrobel, A.G, Benton, D.J, Rosenthal, P.B, Gamblin, S.J. | | Deposit date: | 2020-12-17 | | Release date: | 2020-12-30 | | Last modified: | 2021-02-24 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure and binding properties of Pangolin-CoV spike glycoprotein inform the evolution of SARS-CoV-2.
Nat Commun, 12, 2021
|
|
7BNM
 
 | | Closed conformation of D614G SARS-CoV-2 spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Benton, D.J, Wrobel, A.G, Rosenthal, P.B, Gamblin, S.J. | | Deposit date: | 2021-01-22 | | Release date: | 2021-02-03 | | Last modified: | 2021-03-10 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | The effect of the D614G substitution on the structure of the spike glycoprotein of SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7BNO
 
 | | Open conformation of D614G SARS-CoV-2 spike with 2 Erect RBDs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Benton, D.J, Wrobel, A.G, Rosenthal, P.B, Gamblin, S.J. | | Deposit date: | 2021-01-22 | | Release date: | 2021-02-03 | | Last modified: | 2021-03-10 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | The effect of the D614G substitution on the structure of the spike glycoprotein of SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6N1P
 
 | | Dihedral oligomeric complex of GyrA N-terminal fragment with DNA, solved by cryoEM in C2 symmetry | | Descriptor: | DNA (44-MER), DNA gyrase subunit A | | Authors: | Soczek, K.M, Grant, T, Rosenthal, P.B, Mondragon, A. | | Deposit date: | 2018-11-10 | | Release date: | 2018-12-05 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (6.35 Å) | | Cite: | CryoEM structures of open dimers of Gyrase A in complex with DNA illuminate mechanism of strand passage.
Elife, 7, 2018
|
|
6HKG
 
 | | Structure of FISW84 Fab Fragment | | Descriptor: | FISW84 Fab Fragment Heavy Chain, FISW84 Fab Fragment Light Chain, SULFATE ION | | Authors: | Benton, D.J, Rosenthal, P.B. | | Deposit date: | 2018-09-06 | | Release date: | 2018-09-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Influenza hemagglutinin membrane anchor.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
2OUL
 
 | | The Structure of Chagasin in Complex with a Cysteine Protease Clarifies the Binding Mode and Evolution of a New Inhibitor Family | | Descriptor: | Chagasin, Falcipain 2 | | Authors: | Wang, S.X, Chand, K, Huang, R, Whisstock, J, Jacobelli, J, Fletterick, R.J, Rosenthal, P.J, McKerrow, J.H. | | Deposit date: | 2007-02-11 | | Release date: | 2008-02-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of chagasin in complex with a cysteine protease clarifies the binding mode and evolution of an inhibitor family.
Structure, 15, 2007
|
|
5FO4
 
 | | Crystal structure of the P.falciparum cytosolic leucyl-tRNA synthetase editing domain (space group P1) | | Descriptor: | LEUCYL TRNA SYNTHASE | | Authors: | Palencia, A, Sonoiki, E, Guo, D, Ahyong, V, Dong, C, Li, X, Hernandez, V.S, Gut, J, Legac, J, Cooper, R, Alley, M.R.K, Freund, Y.R, DeRisi, J, Cusack, S, Rosenthal, P.J. | | Deposit date: | 2015-11-18 | | Release date: | 2016-06-22 | | Last modified: | 2016-08-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Anti-Malarial Benzoxaboroles Target P. Falciparum Leucyl-tRNA Synthetase.
Antimicrob.Agents Chemother., 60, 2016
|
|
