3EB5
 
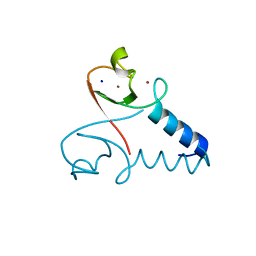 | | Structure of the cIAP2 RING domain | | Descriptor: | Baculoviral IAP repeat-containing protein 3, SODIUM ION, ZINC ION | | Authors: | Mace, P.D, Linke, K, Smith, C.A, Day, C.L. | | Deposit date: | 2008-08-27 | | Release date: | 2008-09-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of the cIAP2 RING domain reveal conformational changes associated with ubiquitin-conjugating enzyme (E2) recruitment.
J.Biol.Chem., 283, 2008
|
|
3EHR
 
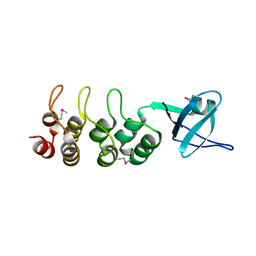 | | Crystal Structure of Human Osteoclast Stimulating Factor | | Descriptor: | Osteoclast-stimulating factor 1 | | Authors: | Tong, S, Zhou, H, Gao, Y, Zhu, Z, Zhang, X, Teng, M, Niu, L. | | Deposit date: | 2008-09-14 | | Release date: | 2009-08-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of human osteoclast stimulating factor
Proteins, 75, 2009
|
|
3EIY
 
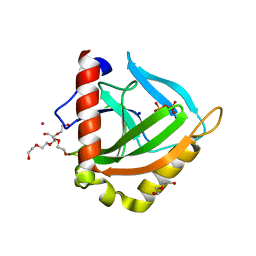 | |
3VQU
 
 | | CRYSTAL STRUCTURE OF HUMAN MPS1 CATALYTIC DOMAIN IN COMPLEX WITH 4-[(4-amino-5-cyano-6-ethoxypyridin-2- yl)amino]benzamide | | Descriptor: | 4-[(4-amino-5-cyano-6-ethoxypyridin-2-yl)amino]benzamide, Dual specificity protein kinase TTK, IODIDE ION | | Authors: | Kusakabe, K, Ide, N, Daigo, Y, Itoh, T, Higashino, K, Okano, Y, Tadano, G, Tachibana, Y, Sato, Y, Inoue, M, Wada, T, Iguchi, M, Kanazawa, T, Ishioka, Y, Dohi, K, Tagashira, S, Kido, Y, Sakamoto, S, Yasuo, K, Maeda, M, Yamamoto, T, Higaki, M, Endoh, T, Ueda, K, Shiota, T, Murai, H, Nakamura, Y. | | Deposit date: | 2012-03-30 | | Release date: | 2012-06-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Diaminopyridine-based potent and selective mps1 kinase inhibitors binding to an unusual flipped-Peptide conformation.
Acs Med.Chem.Lett., 3, 2012
|
|
3ENN
 
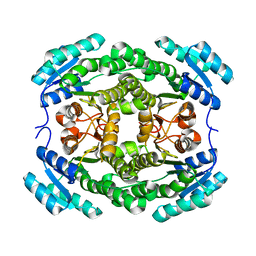 | |
2FNI
 
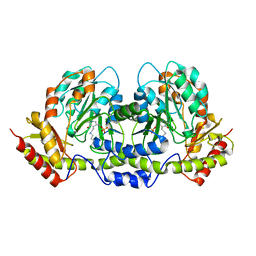 | | PseC aminotransferase involved in pseudoaminic acid biosynthesis | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, aminotransferase | | Authors: | Cygler, M, Matte, A, Lunin, V.V, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2006-01-11 | | Release date: | 2006-01-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and Functional Characterization of PseC, an Aminotransferase Involved in the Biosynthesis of Pseudaminic Acid, an Essential Flagellar Modification in Helicobacter pylori
J.Biol.Chem., 281, 2006
|
|
3V1U
 
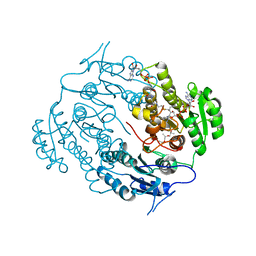 | | Crystal structure of a beta-ketoacyl reductase FabG4 from Mycobacterium tuberculosis H37Rv complexed with NAD+ and Hexanoyl-CoA at 2.5 Angstrom resolution | | Descriptor: | (2S,5R,8R,11S,14S,17S,21R)-5,8,11,14,17-PENTAMETHYL-4,7,10,13,16,19-HEXAOXADOCOSANE-2,21-DIOL, 3-oxoacyl-(Acyl-carrier-protein) reductase, HEXANOYL-COENZYME A, ... | | Authors: | Dutta, D, Bhattacharyya, S, Das, A.K. | | Deposit date: | 2011-12-10 | | Release date: | 2012-11-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of hexanoyl-CoA bound to beta-ketoacyl reductase FabG4 of Mycobacterium tuberculosis
Biochem.J., 450, 2013
|
|
3ERF
 
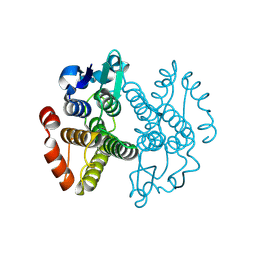 | | Crystal structure of Gtt2 from Saccharomyces cerevisiae | | Descriptor: | Glutathione S-transferase 2 | | Authors: | Ma, X.X, Jiang, Y.L, He, Y.X, Chen, Y.X, Zhou, C.Z. | | Deposit date: | 2008-10-02 | | Release date: | 2009-10-13 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structures of yeast glutathione-S-transferase Gtt2 reveal a new catalytic type of GST family.
Embo Rep., 10, 2009
|
|
3ESK
 
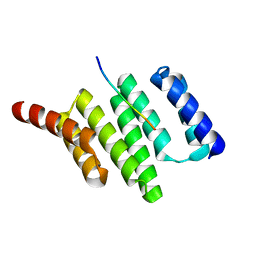 | |
2BD3
 
 | | Porcine pancreatic elastase complexed with beta-casomorphin-7 and Lys-Ala-NH2 at pH 5.0 | | Descriptor: | CALCIUM ION, Chymotrypsin-like elastase family member 1, SULFATE ION, ... | | Authors: | Liu, B, Schofield, C.J, Wilmouth, R.C. | | Deposit date: | 2005-10-20 | | Release date: | 2006-05-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural analyses on intermediates in serine protease catalysis
J.Biol.Chem., 281, 2006
|
|
3E8L
 
 | | The Crystal Structure of the Double-headed Arrowhead Protease Inhibitor A in Complex with Two Trypsins | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Bao, R, Jiang, C.-H, Chi, C.W, Lin, S.X, Chen, Y.X. | | Deposit date: | 2008-08-20 | | Release date: | 2009-07-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | The ternary structure of double-headed arrowhead protease inhibitor API-A complexed with two trypsins reveals a novel reactive site conformation.
J.Biol.Chem., 284, 2009
|
|
3UUL
 
 | |
3UWU
 
 | | Crystal structure of Staphylococcus Aureus triosephosphate isomerase complexed with glycerol-3-phosphate | | Descriptor: | CITRIC ACID, SN-GLYCEROL-3-PHOSPHATE, Triosephosphate isomerase | | Authors: | Mukherjee, S, Roychowdhury, A, Dutta, D, Das, A.K. | | Deposit date: | 2011-12-03 | | Release date: | 2012-10-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structures of triosephosphate isomerase from methicillin resistant Staphylococcus aureus MRSA252 provide structural insights into novel modes of ligand binding and unique conformations of catalytic loop
Biochimie, 94, 2012
|
|
3E5Y
 
 | |
2BD7
 
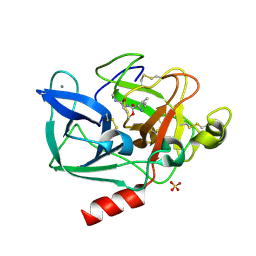 | | Porcine pancreatic elastase complexed with beta-casomorphin-7 and Arg-Phe at pH 5.0 (50 min soak) | | Descriptor: | CALCIUM ION, Chymotrypsin-like elastase family member 1, SULFATE ION, ... | | Authors: | Liu, B, Schofield, C.J, Wilmouth, R.C. | | Deposit date: | 2005-10-20 | | Release date: | 2006-05-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural analyses on intermediates in serine protease catalysis
J.Biol.Chem., 281, 2006
|
|
3SQH
 
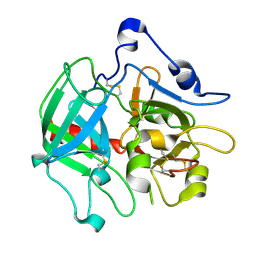 | |
3EPG
 
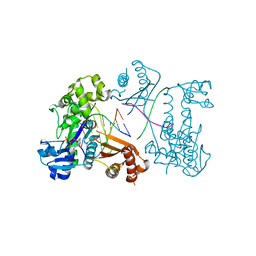 | | Structure of Human DNA Polymerase Iota complexed with N2-ethylguanine | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, 5'-D(*DTP*DCP*DTP*(2EG)P*DGP*DGP*DGP*DTP*DCP*DCP*DTP*DAP*DGP*DGP*DAP*DCP*DCP*(DOC))-3', DNA polymerase iota, ... | | Authors: | Pence, M.G. | | Deposit date: | 2008-09-29 | | Release date: | 2008-12-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Lesion bypass of N2-ethylguanine by human DNA polymerase iota.
J.Biol.Chem., 284, 2009
|
|
3STF
 
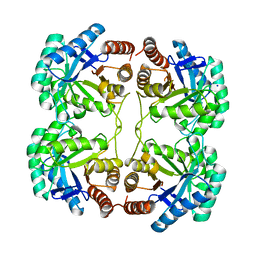 | | Crystal structure of a mutant (S211A) of 3-deoxy-D-manno-octulosonate 8-phosphate synthase (KDO8PS) from Neisseria meningitidis | | Descriptor: | 2-dehydro-3-deoxyphosphooctonate aldolase, CHLORIDE ION, GLYCEROL, ... | | Authors: | Allison, T.M, Jameson, G.B, Gloyne, B.J, Parker, E.J. | | Deposit date: | 2011-07-09 | | Release date: | 2011-11-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | An Extended (beta)7(alpha)7 Substrate-Binding Loop Is Essential for Efficient Catalysis by 3-Deoxy-D-manno-Octulosonate 8-Phosphate Synthase
Biochemistry, 50, 2011
|
|
3EQY
 
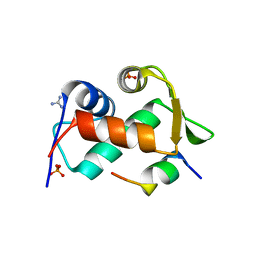 | |
3E4D
 
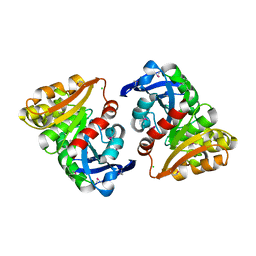 | | Structural and Kinetic Study of an S-Formylglutathione Hydrolase from Agrobacterium tumefaciens | | Descriptor: | CHLORIDE ION, Esterase D, MAGNESIUM ION | | Authors: | Van Straaten, K.E, Gonzalez, C.F, Valladares, R.B, Xu, X, Savchenko, A.V, Sanders, D.A.R. | | Deposit date: | 2008-08-11 | | Release date: | 2009-08-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | The structure of a putative S-formylglutathione hydrolase from Agrobacterium tumefaciens
Protein Sci., 18, 2009
|
|
3VG4
 
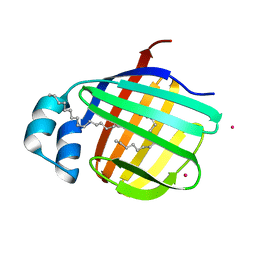 | | Cadmium derivative of human LFABP | | Descriptor: | CADMIUM ION, Fatty acid-binding protein, liver, ... | | Authors: | Sharma, A, Yogavel, M, Sharma, A. | | Deposit date: | 2011-08-03 | | Release date: | 2012-06-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Utility of anion and cation combinations for phasing of protein structures.
J.Struct.Funct.Genom., 13, 2012
|
|
3VZL
 
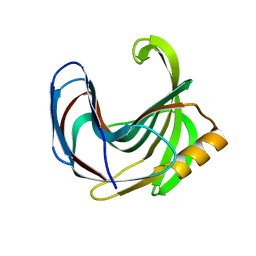 | | Crystal structure of the Bacillus circulans endo-beta-(1,4)-xylanase (BcX) N35H mutant | | Descriptor: | Endo-1,4-beta-xylanase, SULFATE ION | | Authors: | Ludwiczek, M.L, D'Angelo, I, Yalloway, G.N, Okon, M, Nielsen, J.E, Strynadka, N.C, Withers, S.G, McIntosh, L.P. | | Deposit date: | 2012-10-15 | | Release date: | 2013-05-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Strategies for modulating the pH-dependent activity of a family 11 glycoside hydrolase
Biochemistry, 52, 2013
|
|
3EVI
 
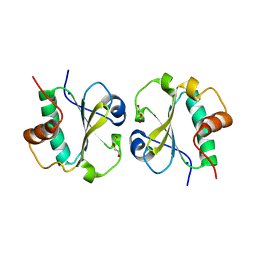 | | Crystal structure of the thioredoxin-fold domain of human phosducin-like protein 2 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Phosducin-like protein 2, SODIUM ION | | Authors: | Lou, X, Bao, R, Zhou, C, Chen, Y. | | Deposit date: | 2008-10-13 | | Release date: | 2009-03-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of the thioredoxin-fold domain of human phosducin-like protein 2
Acta Crystallogr.,Sect.F, 65, 2009
|
|
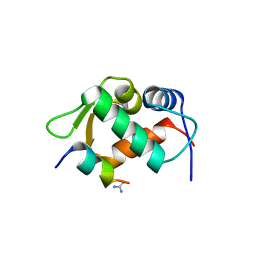
3EQS
 
 | |
2BNO
 
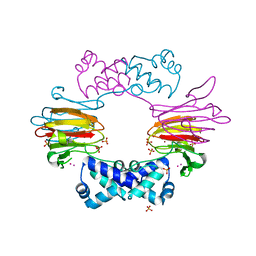 | | The structure of Hydroxypropylphosphonic acid epoxidase from S. wedmorenis. | | Descriptor: | EPOXIDASE, MERCURY (II) ION, SULFATE ION, ... | | Authors: | McLuskey, K, Cameron, S, Hunter, W.N. | | Deposit date: | 2005-03-29 | | Release date: | 2005-10-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and Reactivity of Hydroxypropylphosphonic Acid Epoxidase in Fosfomycin Biosynthesis by a Cation- and Flavin-Dependent Mechanism.
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
