4IAP
 
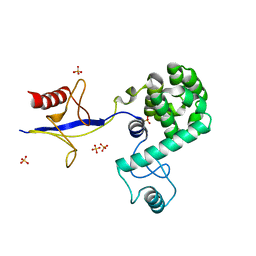 | | Crystal structure of PH domain of Osh3 from Saccharomyces cerevisiae | | Descriptor: | Oxysterol-binding protein homolog 3,Endolysin,Oxysterol-binding protein homolog 3, SULFATE ION | | Authors: | Tong, J, Im, Y.J. | | Deposit date: | 2012-12-07 | | Release date: | 2013-07-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of osh3 reveals a conserved mode of phosphoinositide binding in oxysterol-binding proteins
Structure, 21, 2013
|
|
2Z6B
 
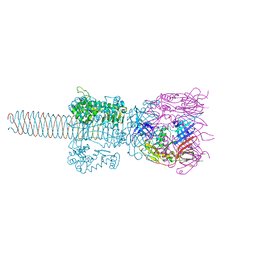 | | Crystal Structure Analysis of (gp27-gp5)3 conjugated with Fe(III) protoporphyrin | | Descriptor: | 1-ETHYL-PYRROLIDINE-2,5-DIONE, Baseplate structural protein Gp27, Tail-associated lysozyme | | Authors: | Koshiyama, T, Yokoi, N, Ueno, T, Kanamaru, S, Nagano, S, Shiro, Y, Arisaka, F, Watanabe, Y. | | Deposit date: | 2007-07-28 | | Release date: | 2008-04-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Molecular design of heteroprotein assemblies providing a bionanocup as a chemical reactor.
Small, 4, 2008
|
|
5YQR
 
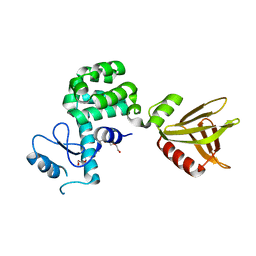 | | Crystal structure of the PH-like domain of Lam6 | | Descriptor: | Endolysin/Membrane-anchored lipid-binding protein LAM6 fusion protein, NONAETHYLENE GLYCOL | | Authors: | Tong, J, Im, Y.J. | | Deposit date: | 2017-11-07 | | Release date: | 2018-01-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.402 Å) | | Cite: | Structural basis of sterol recognition and nonvesicular transport by lipid transfer proteins anchored at membrane contact sites
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5IV5
 
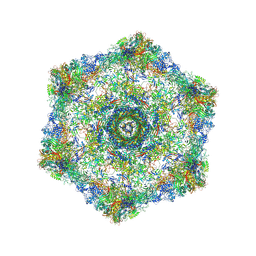 | | Cryo-electron microscopy structure of the hexagonal pre-attachment T4 baseplate-tail tube complex | | Descriptor: | Baseplate hub protein gp27, Baseplate tail-tube protein gp48, Baseplate tail-tube protein gp54, ... | | Authors: | Taylor, N.M.I, Guerrero-Ferreira, R.C, Goldie, K.N, Stahlberg, H, Leiman, P.G. | | Deposit date: | 2016-03-19 | | Release date: | 2016-05-18 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.11 Å) | | Cite: | Structure of the T4 baseplate and its function in triggering sheath contraction.
Nature, 533, 2016
|
|
7MI8
 
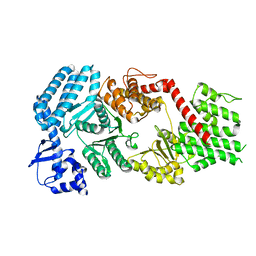 | | Signal subtracted reconstruction of AAA5 and AAA6 domains of dynein in the presence of a pyrazolo-pyrimidinone-based compound, Model 5 | | Descriptor: | Fusion protein of Dynein and Endolysin | | Authors: | Santarossa, C.C, Coudray, N, Urnavicius, L, Ekiert, D.C, Bhabha, G, Kapoor, T.M. | | Deposit date: | 2021-04-16 | | Release date: | 2021-05-26 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Targeting allostery in the Dynein motor domain with small molecule inhibitors.
Cell Chem Biol, 28, 2021
|
|
7MI6
 
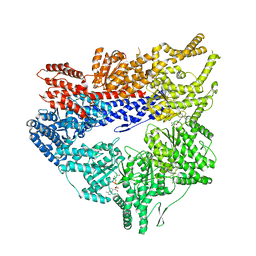 | | Yeast dynein motor domain in the presence of a pyrazolo-pyrimidinone-based compound, Model 1 | | Descriptor: | (8S)-6-(3-bromophenoxy)-2-[1-(4-chlorophenyl)cyclopropyl]-7-hydroxypyrazolo[1,5-a]pyrimidine-3-carbonitrile, ADENOSINE-5'-TRIPHOSPHATE, Fusion protein of Dynein and Endolysin, ... | | Authors: | Santarossa, C.C, Urnavicius, L, Coudray, N, Ekeirt, D.C, Bhabha, G, Kapoor, T.M. | | Deposit date: | 2021-04-16 | | Release date: | 2021-05-26 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Targeting allostery in the Dynein motor domain with small molecule inhibitors.
Cell Chem Biol, 28, 2021
|
|
7MI3
 
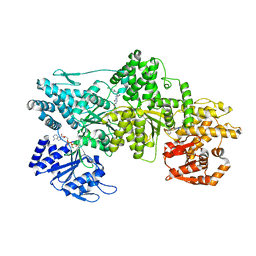 | | Signal subtracted reconstruction of AAA2, AAA3, and AAA4 domains of dynein in the presence of a pyrazolo-pyrimidinone-based compound, Model 4 | | Descriptor: | (8S)-6-(3-bromophenoxy)-2-[1-(4-chlorophenyl)cyclopropyl]-7-hydroxypyrazolo[1,5-a]pyrimidine-3-carbonitrile, ADENOSINE-5'-TRIPHOSPHATE, Fusion protein of Dynein and Endolysin, ... | | Authors: | Santarossa, C.C, Coudray, N, Urnavicius, L, Ekiert, D.C, Bhabha, G, Kapoor, T.M. | | Deposit date: | 2021-04-16 | | Release date: | 2021-05-26 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Targeting allostery in the Dynein motor domain with small molecule inhibitors.
Cell Chem Biol, 28, 2021
|
|
7MI1
 
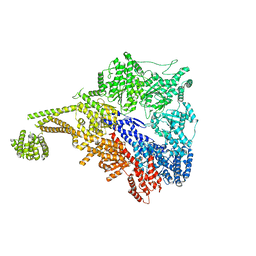 | |
2QB0
 
 | |
2QAR
 
 | |
1PDL
 
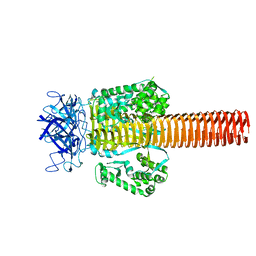 | | Fitting of gp5 in the cryoEM reconstruction of the bacteriophage T4 baseplate | | Descriptor: | Tail-associated lysozyme | | Authors: | Kostyuchenko, V.A, Leiman, P.G, Chipman, P.R, Kanamaru, S, van Raaij, M.J, Arisaka, F, Mesyanzhinov, V.V, Rossmann, M.G. | | Deposit date: | 2003-05-19 | | Release date: | 2003-09-09 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Three-dimensional structure of the bacteriophage T4 baseplate
Nat.Struct.Biol., 10, 2003
|
|
1K28
 
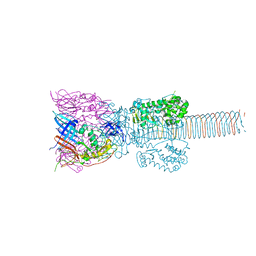 | | The Structure of the Bacteriophage T4 Cell-Puncturing Device | | Descriptor: | BASEPLATE STRUCTURAL PROTEIN GP27, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Kanamaru, S, Leiman, P.G, Kostyuchenko, V.A, Chipman, P.R, Mesyanzhinov, V.V, Arisaka, F, Rossmann, M.G. | | Deposit date: | 2001-09-26 | | Release date: | 2002-02-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of the cell-puncturing device of bacteriophage T4.
Nature, 415, 2002
|
|
6WJC
 
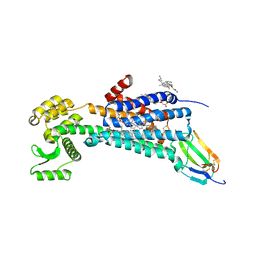 | | Muscarinic acetylcholine receptor 1 - muscarinic toxin 7 complex | | Descriptor: | (1R,5S)-8-METHYL-8-AZABICYCLO[3.2.1]OCT-3-YL (2R)-3-HYDROXY-2-PHENYLPROPANOATE, ACETAMIDE, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Maeda, S, Kobilka, B.K. | | Deposit date: | 2020-04-13 | | Release date: | 2020-07-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure and selectivity engineering of the M1muscarinic receptor toxin complex.
Science, 369, 2020
|
|
3D4S
 
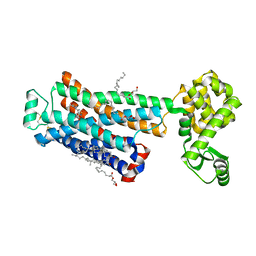 | | Cholesterol bound form of human beta2 adrenergic receptor. | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-1-(tert-butylamino)-3-[(4-morpholin-4-yl-1,2,5-thiadiazol-3-yl)oxy]propan-2-ol, Beta-2 adrenergic receptor/T4-lysozyme chimera, ... | | Authors: | Hanson, M.A, Cherezov, V, Roth, C.B, Griffith, M.T, Jaakola, V.-P, Chien, E.Y.T, Velasquez, J, Kuhn, P, Stevens, R.C, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D), GPCR Network (GPCR) | | Deposit date: | 2008-05-14 | | Release date: | 2008-06-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A specific cholesterol binding site is established by the 2.8 A structure of the human beta2-adrenergic receptor.
Structure, 16, 2008
|
|
7SYF
 
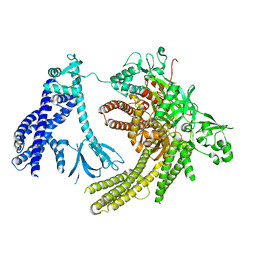 | | Reconstruction of full-length Prex-1 (PtdIns(3,4,5)P3-dependent Rac Exchanger 1) | | Descriptor: | Phosphatidylinositol 3,4,5-trisphosphate-dependent Rac exchanger 1 protein,Endolysin chimera | | Authors: | Lupton, C.J, Bayly-Jones, C, Ellisdon, A.M. | | Deposit date: | 2021-11-24 | | Release date: | 2022-07-27 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structure of the metastatic factor P-Rex1 reveals a two-layered autoinhibitory mechanism.
Nat.Struct.Mol.Biol., 29, 2022
|
|
4PLA
 
 | |
7RX9
 
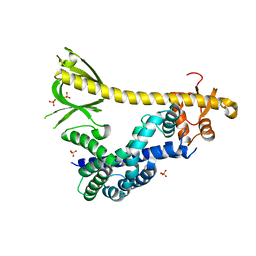 | | Structure of autoinhibited P-Rex1 | | Descriptor: | Phosphatidylinositol 3,4,5-trisphosphate-dependent Rac exchanger 1 protein, Endolysin chimera, SULFATE ION | | Authors: | Ellisdon, A.M, Chang, Y. | | Deposit date: | 2021-08-22 | | Release date: | 2022-08-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | Structure of the metastatic factor P-Rex1 reveals a two-layered autoinhibitory mechanism.
Nat.Struct.Mol.Biol., 29, 2022
|
|
4U16
 
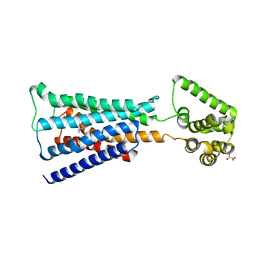 | | M3-mT4L receptor bound to NMS | | Descriptor: | D(-)-TARTARIC ACID, Muscarinic acetylcholine receptor M3,Lysozyme,Muscarinic acetylcholine receptor M3, N-methyl scopolamine | | Authors: | Thorsen, T.S, Matt, R, Weis, W.I, Kobilka, B. | | Deposit date: | 2014-07-15 | | Release date: | 2014-11-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Modified T4 Lysozyme Fusion Proteins Facilitate G Protein-Coupled Receptor Crystallogenesis.
Structure, 22, 2014
|
|
6QAJ
 
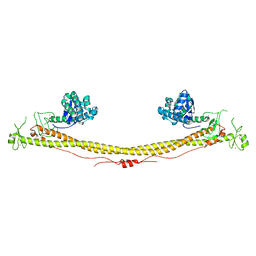 | | Structure of the tripartite motif of KAP1/TRIM28 | | Descriptor: | Endolysin,Transcription intermediary factor 1-beta, ZINC ION | | Authors: | Stoll, G.A, Oda, S, Yu, M, Modis, Y. | | Deposit date: | 2018-12-19 | | Release date: | 2019-07-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.901 Å) | | Cite: | Structure of KAP1 tripartite motif identifies molecular interfaces required for retroelement silencing.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
7P2L
 
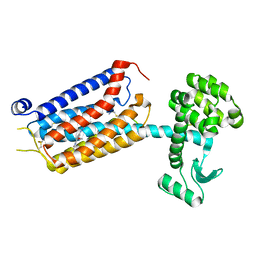 | | thermostabilised 7TM domain of human mGlu5 receptor bound to photoswitchable ligand alloswitch-1 | | Descriptor: | 2-chloranyl-~{N}-[2-methoxy-4-[(~{E})-pyridin-2-yldiazenyl]phenyl]benzamide, Metabotropic glutamate receptor 5,Endolysin,Metabotropic glutamate receptor 5 | | Authors: | Huang, C.Y, Vinothkumar, K.R, Lebon, G. | | Deposit date: | 2021-07-06 | | Release date: | 2021-09-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Agonists and allosteric modulators promote signaling from different metabotropic glutamate receptor 5 conformations.
Cell Rep, 36, 2021
|
|
6PS5
 
 | | XFEL beta2 AR structure by ligand exchange from Timolol to Propranolol. | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 1-(ISOPROPYLAMINO)-3-(1-NAPHTHYLOXY)-2-PROPANOL, CHOLESTEROL, ... | | Authors: | Ishchenko, A, Stauch, B, Han, G.W, Batyuk, A, Shiriaeva, A, Li, C, Zatsepin, N.A, Weierstall, U, Liu, W, Nango, E, Nakane, T, Tanaka, R, Tono, K, Joti, Y, Iwata, S, Moraes, I, Gati, C, Cherezov, C. | | Deposit date: | 2019-07-12 | | Release date: | 2019-11-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Toward G protein-coupled receptor structure-based drug design using X-ray lasers.
Iucrj, 6, 2019
|
|
3VW7
 
 | | Crystal structure of human protease-activated receptor 1 (PAR1) bound with antagonist vorapaxar at 2.2 angstrom | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHLORIDE ION, Proteinase-activated receptor 1, ... | | Authors: | Zhang, C, Srinivasan, Y, Arlow, D.H, Fung, J.J, Palmer, D, Zheng, Y, Green, H.F, Pandey, A, Dror, R.O, Shaw, D.E, Weis, W.I, Coughlin, S.R, Kobilka, B.K. | | Deposit date: | 2012-08-07 | | Release date: | 2012-12-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | High-resolution crystal structure of human protease-activated receptor 1
Nature, 492, 2012
|
|
8A5X
 
 | |
8JWY
 
 | | Crystal structure of A2AR-T4L in complex with 2-118 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 3-[2-azanyl-6-[2-oxidanylidene-1-[[6-(2-oxidanylpropan-2-yl)pyridin-2-yl]methyl]pyridin-4-yl]pyrimidin-4-yl]-2-methyl-benzenecarbonitrile, Adenosine receptor A2a,Endolysin, ... | | Authors: | Weng, Y, Chen, Y, Xu, Y, Song, G. | | Deposit date: | 2023-06-29 | | Release date: | 2023-08-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structural insight into the dual-antagonistic mechanism of AB928 on adenosine A 2 receptors.
Sci China Life Sci, 67, 2024
|
|
8JWZ
 
 | | Crystal structure of A2AR-T4L in complex with AB928 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 3-[2-azanyl-6-[1-[[6-(2-oxidanylpropan-2-yl)pyridin-2-yl]methyl]-1,2,3-triazol-4-yl]pyrimidin-4-yl]-2-methyl-benzenecarbonitrile, Adenosine receptor A2a,Endolysin, ... | | Authors: | Weng, Y, Chen, Y, Xu, Y, Song, G. | | Deposit date: | 2023-06-29 | | Release date: | 2023-08-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structural insight into the dual-antagonistic mechanism of AB928 on adenosine A 2 receptors.
Sci China Life Sci, 67, 2024
|
|
