4PI3
 
 | |
4R5H
 
 | | Crystal structure of sp-Aspartate-Semialdehyde-Dehydrogenase with Nicotinamide-Adenine-Dinucleotide-Phosphate and 3-carboxy-propenyl-phthalic acid | | Descriptor: | 1,2-ETHANEDIOL, 3-[(1E)-3-carboxyprop-1-en-1-yl]benzene-1,2-dicarboxylic acid, Aspartate-semialdehyde dehydrogenase, ... | | Authors: | Pavlovsky, A.G, Thangavelu, B, Bhansali, P, Viola, R.E. | | Deposit date: | 2014-08-21 | | Release date: | 2014-12-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A cautionary tale of structure-guided inhibitor development against an essential enzyme in the aspartate-biosynthetic pathway.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4NTI
 
 | | Crystal structure of D60N mutant of Arabidopsis ACD11 (accelerated-cell-death 11) complexed with C12 ceramide-1-phosphate (d18:1/12:0) at 2.9 Angstrom resolution | | Descriptor: | (2S,3R,4E)-2-(dodecanoylamino)-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, DI(HYDROXYETHYL)ETHER, accelerated-cell-death 11 | | Authors: | Simanshu, D.K, Brown, R.E, Patel, D.J. | | Deposit date: | 2013-12-02 | | Release date: | 2014-02-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.899 Å) | | Cite: | Arabidopsis Accelerated Cell Death 11, ACD11, Is a Ceramide-1-Phosphate Transfer Protein and Intermediary Regulator of Phytoceramide Levels.
Cell Rep, 6, 2014
|
|
4NTG
 
 | | Crystal structure of D60A mutant of Arabidopsis ACD11 (accelerated-cell-death 11) complexed with C12 ceramide-1-phosphate (d18:1/12:0) at 2.55 Angstrom resolution | | Descriptor: | (2S,3R,4E)-2-(dodecanoylamino)-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, accelerated-cell-death 11 | | Authors: | Simanshu, D.K, Brown, R.E, Patel, D.J. | | Deposit date: | 2013-12-02 | | Release date: | 2014-02-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5505 Å) | | Cite: | Arabidopsis Accelerated Cell Death 11, ACD11, Is a Ceramide-1-Phosphate Transfer Protein and Intermediary Regulator of Phytoceramide Levels.
Cell Rep, 6, 2014
|
|
4R68
 
 | | Lactate Dehydrogenase in complex with inhibitor compound 31 | | Descriptor: | (1S)-1-phenylethyl (4-chloro-3-{[(4S)-4-(2,6-dichlorophenyl)-2-hydroxy-6-oxocyclohex-1-en-1-yl]sulfanyl}phenyl)acetate, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Eigenbrot, C, Ultsch, M. | | Deposit date: | 2014-08-22 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.112 Å) | | Cite: | Optimization of 5-(2,6-dichlorophenyl)-3-hydroxy-2-mercaptocyclohex-2-enones as potent inhibitors of human lactate dehydrogenase.
Bioorg.Med.Chem.Lett., 25, 2014
|
|
4O5T
 
 | | Crystal structure of Diels-Alderase CE20 in complex with a product analog | | Descriptor: | 4-{[2-(phosphonooxy)ethyl]carbamoyl}benzyl [(1R,6S)-6-(dimethylcarbamoyl)cyclohex-2-en-1-yl]carbamate, Diisopropyl-fluorophosphatase | | Authors: | Beck, T, Preiswerk, N, Mayer, C, Hilvert, D. | | Deposit date: | 2013-12-20 | | Release date: | 2014-06-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Impact of scaffold rigidity on the design and evolution of an artificial Diels-Alderase.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4OBV
 
 | | Ruminococcus gnavus tryptophan decarboxylase RUMGNA_01526 (alpha-FMT) | | Descriptor: | Pyridoxal-dependent decarboxylase domain protein, alpha-(fluoromethyl)-D-tryptophan, {5-hydroxy-4-[(1E)-4-(1H-indol-3-yl)-3-oxobut-1-en-1-yl]-6-methylpyridin-3-yl}methyl dihydrogen phosphate | | Authors: | Fraser, J.S, Van Benschoten, A.H. | | Deposit date: | 2014-01-07 | | Release date: | 2014-10-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Discovery and Characterization of Gut Microbiota Decarboxylases that Can Produce the Neurotransmitter Tryptamine.
Cell Host Microbe, 16, 2014
|
|
4PV1
 
 | | Cytochrome B6F structure from M. laminosus with the quinone analog inhibitor stigmatellin | | Descriptor: | (1R)-2-(dodecanoyloxy)-1-[(phosphonooxy)methyl]ethyl tetradecanoate, (7R,17E)-4-HYDROXY-N,N,N,7-TETRAMETHYL-7-[(8E)-OCTADEC-8-ENOYLOXY]-10-OXO-3,5,9-TRIOXA-4-PHOSPHAHEPTACOS-17-EN-1-AMINIUM 4-OXIDE, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, ... | | Authors: | Hasan, S.S, Yamashita, E, Cramer, W.A. | | Deposit date: | 2014-03-14 | | Release date: | 2014-08-20 | | Last modified: | 2021-03-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Traffic within the cytochrome b6f lipoprotein complex: gating of the quinone portal.
Biophys.J., 107, 2014
|
|
4PV5
 
 | | Crystal structure of mouse glyoxalase I in complexed with 18-beta-glycyrrhetinic acid | | Descriptor: | (3BETA,5BETA,14BETA)-3-HYDROXY-11-OXOOLEAN-12-EN-29-OIC ACID, Lactoylglutathione lyase, ZINC ION | | Authors: | Zhang, H, Zhai, J, Zhang, L.P, Zhao, Y.N, Li, C, Hu, X.P. | | Deposit date: | 2014-03-15 | | Release date: | 2015-03-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for 18-beta-glycyrrhetinic acid as a novel non-GSH analog glyoxalase I inhibitor
Acta Pharmacol.Sin., 36, 2015
|
|
4Q0O
 
 | | Crystal Structure of the fifth bromodomain of Human Poly-bromodomain containing protein 1 (PB1) in complex with a hydroxyphenyl-propenone ligand | | Descriptor: | (2E)-1-(2-hydroxyphenyl)-3-[(3R)-3-phenylpiperidin-1-yl]prop-2-en-1-one, POTASSIUM ION, Protein polybromo-1 | | Authors: | Filippakopoulos, P, Picaud, S, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-04-02 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal Structure of the fifth bromodomain of Human Poly-bromodomain containing protein 1 (PB1) in complex with a hydroxyphenyl-propenone ligand
To be Published
|
|
4QNQ
 
 | |
4R69
 
 | | Lactate Dehydrogenase in complex with inhibitor compound 13 | | Descriptor: | (5R)-2-[(2-chlorophenyl)sulfanyl]-5-[2,6-dichloro-3-(tetrahydro-2H-pyran-4-ylamino)phenyl]-3-hydroxycyclohex-2-en-1-one, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Eigenbrot, C, Ultsch, M. | | Deposit date: | 2014-08-22 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Optimization of 5-(2,6-dichlorophenyl)-3-hydroxy-2-mercaptocyclohex-2-enones as potent inhibitors of human lactate dehydrogenase.
Bioorg.Med.Chem.Lett., 25, 2014
|
|
4Q2F
 
 | | Galectin-1 in Complex with Ligand AN020 | | Descriptor: | Galectin-1, SULFATE ION, prop-2-en-1-yl 2-(acetylamino)-4-O-(3-O-{[1-(5-amino-1H-1,2,4-triazol-3-yl)-1H-1,2,3-triazol-4-yl]methyl}-beta-D-galactopyranosyl)-2-deoxy-beta-D-glucopyranoside | | Authors: | Grimm, C, Bertleff-Zieschang, N. | | Deposit date: | 2014-04-08 | | Release date: | 2015-10-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Galectin-1 in Complex with Ligand AN020
To be Published
|
|
4QO8
 
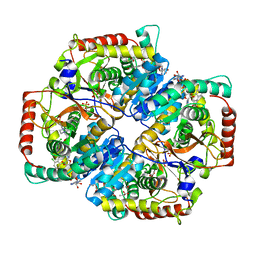 | | Lactate Dehydrogenase A in complex with substituted 3-Hydroxy-2-mercaptocyclohex-2-enone compound 104 | | Descriptor: | (5S)-2-[(2-chlorophenyl)sulfanyl]-5-(2,6-dichlorophenyl)-3-hydroxycyclohex-2-en-1-one, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Eigenbrot, C, Ultsch, M. | | Deposit date: | 2014-06-19 | | Release date: | 2014-07-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Identification of substituted 3-hydroxy-2-mercaptocyclohex-2-enones as potent inhibitors of human lactate dehydrogenase.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
4R3B
 
 | |
4RN2
 
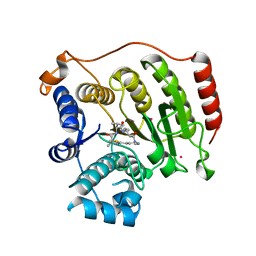 | | Crystal structure of S39D HDAC8 in complex with a largazole analogue. | | Descriptor: | (5R,8S,11S)-5-methyl-8-(propan-2-yl)-11-[(1E)-4-sulfanylbut-1-en-1-yl]-3-thia-7,10,14,17,21-pentaazatricyclo[14.3.1.1~2,5~]henicosa-1(20),2(21),16,18-tetraene-6,9,13-trione, Histone deacetylase 8, POTASSIUM ION, ... | | Authors: | Decroos, C, Christianson, D.W. | | Deposit date: | 2014-10-22 | | Release date: | 2015-04-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Variable Active Site Loop Conformations Accommodate the Binding of Macrocyclic Largazole Analogues to HDAC8.
Biochemistry, 54, 2015
|
|
4RLW
 
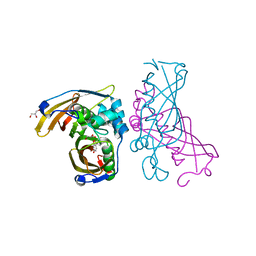 | | Crystal Structure of (3R)-hydroxyacyl-ACP dehydratase HadAB hetero-dimer from Mycobacterium tuberculosis complexed with Butein | | Descriptor: | (2E)-1-(2,4-dihydroxyphenyl)-3-(3,4-dihydroxyphenyl)prop-2-en-1-one, (3R)-hydroxyacyl-ACP dehydratase subunit HadA, (3R)-hydroxyacyl-ACP dehydratase subunit HadB, ... | | Authors: | Li, J, Dong, Y, Rao, Z.H. | | Deposit date: | 2014-10-18 | | Release date: | 2015-10-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.196 Å) | | Cite: | Molecular basis for the inhibition of beta-hydroxyacyl-ACP dehydratase HadAB complex from Mycobacterium tuberculosis by flavonoid inhibitors.
Protein Cell, 6, 2015
|
|
4OIC
 
 | | Crystal structrual of a soluble protein | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Bet v I allergen-like, CHLORIDE ION, ... | | Authors: | He, Y, Hao, Q, Li, W, Yan, C, Yan, N, Yin, P. | | Deposit date: | 2014-01-19 | | Release date: | 2015-01-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Identification and characterization of ABA receptors in Oryza sativa
Plos One, 9, 2014
|
|
4RSE
 
 | | Crystal structure of RPE65 in complex with MB-001 and palmitate | | Descriptor: | (1R)-3-amino-1-{3-[(2,6,6-trimethylcyclohex-1-en-1-yl)methoxy]phenyl}propan-1-ol, FE (II) ION, PALMITIC ACID, ... | | Authors: | Kiser, P.D, Shi, W, Palczewski, K. | | Deposit date: | 2014-11-07 | | Release date: | 2015-04-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Catalytic mechanism of a retinoid isomerase essential for vertebrate vision.
Nat.Chem.Biol., 11, 2015
|
|
4RME
 
 | | Crystal structure of human Retinoid X receptor alpha ligand binding domain complex with 9cUAB111 and coactivator peptide GRIP-1 | | Descriptor: | (2E,4E,6Z,8E)-3,7-dimethyl-8-[2-(3-methylbutyl)-3-(propan-2-yl)cyclohex-2-en-1-ylidene]octa-2,4,6-trienoic acid, Nuclear receptor coactivator 2, Retinoic acid receptor RXR-alpha | | Authors: | Xia, G, Muccio, D.D. | | Deposit date: | 2014-10-21 | | Release date: | 2015-09-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Conformationally Defined Rexinoids and Their Efficacy in the Prevention of Mammary Cancers.
J.Med.Chem., 58, 2015
|
|
4RN1
 
 | | Crystal structure of S39D HDAC8 in complex with a largazole analogue. | | Descriptor: | (5R,8S,11S)-5-methyl-8-(propan-2-yl)-11-[(1E)-4-sulfanylbut-1-en-1-yl]-3-thia-7,10,14,20,21-pentaazatricyclo[14.3.1.1~2,5~]henicosa-1(20),2(21),16,18-tetraene-6,9,13-trione, GLYCEROL, Histone deacetylase 8, ... | | Authors: | Decroos, C, Christianson, D.W. | | Deposit date: | 2014-10-22 | | Release date: | 2015-04-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Variable Active Site Loop Conformations Accommodate the Binding of Macrocyclic Largazole Analogues to HDAC8.
Biochemistry, 54, 2015
|
|
8CLV
 
 | |
8CLQ
 
 | |
8CMO
 
 | | Cryo-EM structure of the Photosystem I - LHCI supercomplex from Coelastrella sp. | | Descriptor: | (1~{S})-3,5,5-trimethyl-4-[(1~{E},3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-3,7,12,16-tetramethyl-18-[(4~{S})-2,6,6-trimethyl-4-oxidanyl-cyclohexen-1-yl]octadeca-1,3,5,7,9,11,13,15,17-nonaenyl]cyclohex-3-en-1-ol, (2R)-2-hydroxy-3-(phosphonooxy)propyl (9E)-octadec-9-enoate, (2S)-3-{[(R)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-hydroxypropyl hexadecanoate, ... | | Authors: | Fadeeva, M, Klaiman, D, Nelson, N. | | Deposit date: | 2023-02-20 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Cryo-EM structure of the Photosystem I - LHCI supercomplex from Coelastrella sp.
To Be Published
|
|
4XUI
 
 | |
