1S5N
 
 | | Xylose Isomerase in Substrate and Inhibitor Michaelis States: Atomic Resolution Studies of a Metal-Mediated Hydride Shift | | Descriptor: | HYDROXIDE ION, MANGANESE (II) ION, SODIUM ION, ... | | Authors: | Fenn, T.D, Ringe, D, Petsko, G.A. | | Deposit date: | 2004-01-21 | | Release date: | 2004-02-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Xylose isomerase in substrate and inhibitor michaelis States: atomic resolution studies of a metal-mediated hydride shift(,).
Biochemistry, 43, 2004
|
|
6Y0Y
 
 | | Sarcin Ricin Loop, mutant C2666U | | Descriptor: | MAGNESIUM ION, RNA (27-MER), SODIUM ION | | Authors: | Ennifar, E, Westhof, E. | | Deposit date: | 2020-02-10 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Sarcin Ricin Loop, mutant C2666U
To Be Published
|
|
3NJ6
 
 | | 0.95 A resolution X-ray structure of (GGCAGCAGCC)2 | | Descriptor: | 5'-R(*GP*GP*CP*AP*GP*CP*AP*GP*CP*C)-3', SULFATE ION | | Authors: | Kiliszek, A, Kierzek, R, Krzyzosiak, W.J, Rypniewski, W. | | Deposit date: | 2010-06-17 | | Release date: | 2010-08-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Atomic resolution structure of CAG RNA repeats: structural insights and implications for the trinucleotide repeat expansion diseases.
Nucleic Acids Res., 38, 2010
|
|
5AQ0
 
 | | The structure of the Transthyretin-like domain of the first catalytic domain of the HUMAN Carboxypeptidase D | | Descriptor: | CARBOXYPEPTIDASE D, GLYCEROL | | Authors: | Gallego, P, Garcia-Pardo, J, Lorenzo, J, Aviles, F.X, Ventura, S, Reverter, D. | | Deposit date: | 2015-09-18 | | Release date: | 2016-09-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | The Structure of the Ttldomain of the Human Carboxypeptidase D
To be Published
|
|
7QQC
 
 | | Structure of CTX-M-15 K73A mutant | | Descriptor: | Beta-lactamase, CHLORIDE ION, SULFATE ION | | Authors: | Tooke, C.L, Hinchliffe, P, Spencer, J. | | Deposit date: | 2022-01-07 | | Release date: | 2022-05-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Penicillanic Acid Sulfones Inactivate the Extended-Spectrum beta-Lactamase CTX-M-15 through Formation of a Serine-Lysine Cross-Link: an Alternative Mechanism of beta-Lactamase Inhibition.
Mbio, 13, 2022
|
|
6FO5
 
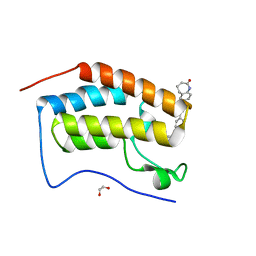 | | FIRST DOMAIN OF HUMAN BROMODOMAIN BRD4 IN COMPLEX WITH INHIBITOR #17 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, ~{N}-[[4-[[7-ethyl-2,6-bis(oxidanylidene)purin-3-yl]methyl]phenyl]methyl]-2-oxidanylidene-1,3,4,5-tetrahydro-1-benzazepine-7-sulfonamide | | Authors: | Raux, B, Betzi, S. | | Deposit date: | 2018-02-06 | | Release date: | 2018-06-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Integrated Strategy for Lead Optimization Based on Fragment Growing: The Diversity-Oriented-Target-Focused-Synthesis Approach.
J. Med. Chem., 61, 2018
|
|
7QQ5
 
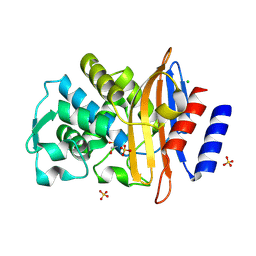 | |
1MXT
 
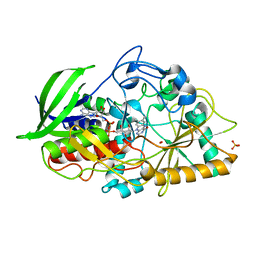 | | Atomic resolution structure of Cholesterol oxidase (Streptomyces sp. SA-COO) | | Descriptor: | CHOLESTEROL OXIDASE, FLAVIN-N7 PROTONATED-ADENINE DINUCLEOTIDE, OXYGEN MOLECULE, ... | | Authors: | Vrielink, A, Lario, P.I. | | Deposit date: | 2002-10-03 | | Release date: | 2003-02-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Sub-atomic resolution crystal structure of cholesterol oxidase:
What atomic resolution crystallography reveals about enzyme mechanism and the role of FAD cofactor in redox activity
J.Mol.Biol., 326, 2003
|
|
7JX4
 
 | | Crystal Structure of N-Lysine Peptoid-modified Collagen Triple Helix | | Descriptor: | Collagen mimetic peptide with N-Lysine guest | | Authors: | Yu, M.S, Whitby, F.G, Hill, C.P, Kessler, J.L, Yang, L.D. | | Deposit date: | 2020-08-26 | | Release date: | 2021-07-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Peptoid Residues Make Diverse, Hyperstable Collagen Triple-Helices.
J.Am.Chem.Soc., 143, 2021
|
|
3NED
 
 | | mRouge | | Descriptor: | ACETATE ION, PAmCherry1 protein, SODIUM ION | | Authors: | Mayo, S.L, Chica, R.A, Moore, M.M. | | Deposit date: | 2010-06-08 | | Release date: | 2010-11-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Generation of longer emission wavelength red fluorescent proteins using computationally designed libraries.
Proc. Natl. Acad. Sci. U.S.A., 107, 2010
|
|
2ZQA
 
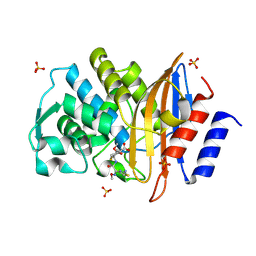 | | Cefotaxime acyl-intermediate structure of class a beta-lacta Toho-1 E166A/R274N/R276N triple mutant | | Descriptor: | Beta-lactamase Toho-1, CEFOTAXIME, C3' cleaved, ... | | Authors: | Nitanai, Y, Shimamura, T, Uchiyama, T, Ishii, Y, Takehira, M, Yutani, K, Matsuzawa, H, Miyano, M. | | Deposit date: | 2008-08-07 | | Release date: | 2009-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Structural Basis of Extend Spectrum Beta-Lactamase in Correlation of Enzymatic Kinetics and Thermal Stability of Acyl-Intermediates
To be Published
|
|
6TJ9
 
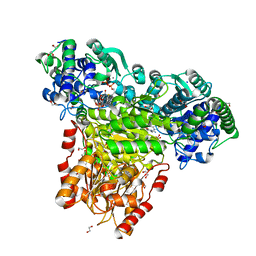 | | Escherichia coli transketolase in complex with cofactor analog 2'-methoxythiamine and substrate xylulose 5-phosphate | | Descriptor: | 1,2-ETHANEDIOL, 2-[3-[(4-azanyl-2-methoxy-pyrimidin-5-yl)methyl]-4-methyl-1,3-thiazol-5-yl]ethyl phosphono hydrogen phosphate, 5-O-phosphono-D-xylulose, ... | | Authors: | Rabe von Pappenheim, F, Tittmann, K. | | Deposit date: | 2019-11-25 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Structural basis for antibiotic action of the B 1 antivitamin 2'-methoxy-thiamine.
Nat.Chem.Biol., 16, 2020
|
|
4QIO
 
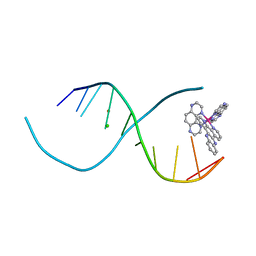 | | Lambda-[Ru(TAP)2(dppz)]2+ bound to d(TCGGCGCCIA) at high resolution | | Descriptor: | 5'-D(*TP*CP*GP*GP*CP*GP*CP*CP*IP*A)-3', BARIUM ION, CHLORIDE ION, ... | | Authors: | Gurung, S.P, Hall, J.P, Cardin, C.J. | | Deposit date: | 2014-06-01 | | Release date: | 2015-06-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Inosine Can Increase DNA's Susceptibility to Photo-oxidation by a Ru II Complex due to Structural Change in the Minor Groove.
Chemistry, 23, 2017
|
|
1RTQ
 
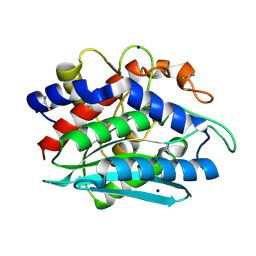 | | The 0.95 Angstrom Resolution Crystal Structure of the Aminopeptidase from Aeromonas proteolytica | | Descriptor: | Bacterial leucyl aminopeptidase, SODIUM ION, THIOCYANATE ION, ... | | Authors: | Desmarais, W, Bienvenue, D.L, Krzysztof, B.P, Holz, R.C, Petsko, G.A, Ringe, D. | | Deposit date: | 2003-12-10 | | Release date: | 2004-02-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | The high-resolution structures of the neutral and the low pH crystals of aminopeptidase from Aeromonas proteolytica.
J.Biol.Inorg.Chem., 11, 2006
|
|
5JSK
 
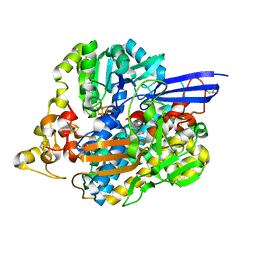 | | The 3D structure of [NiFeSe] hydrogenase from Desulfovibrio vulgaris Hildenborough in the reduced state at 0.95 Angstrom resolution | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE (II) ION, GLYCEROL, ... | | Authors: | Marques, M.C, Pereira, I.A.C, Matias, P.M. | | Deposit date: | 2016-05-08 | | Release date: | 2017-03-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | The direct role of selenocysteine in [NiFeSe] hydrogenase maturation and catalysis.
Nat. Chem. Biol., 13, 2017
|
|
6WEY
 
 | |
5OAV
 
 | |
5XP6
 
 | | native structure of NDM-1 crystallized at pH5.5 | | Descriptor: | 1,2-ETHANEDIOL, HYDROXIDE ION, Metallo-beta-lactamase type 2, ... | | Authors: | Zhang, H, Ma, G, Lai, J, Sun, H. | | Deposit date: | 2017-06-01 | | Release date: | 2018-03-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Bismuth antimicrobial drugs serve as broad-spectrum metallo-beta-lactamase inhibitors
Nat Commun, 9, 2018
|
|
6YZN
 
 | | Closo-carborane butyl-sulfonamide in complex with CA IX mimic | | Descriptor: | Carbonic anhydrase 2, Carborane closo-butyl-sulfonamide, ZINC ION | | Authors: | Kugler, M, Brynda, J, Pospisilova, K, Rezacova, P. | | Deposit date: | 2020-05-07 | | Release date: | 2020-10-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | The structural basis for the selectivity of sulfonamido dicarbaboranes toward cancer-associated carbonic anhydrase IX.
J Enzyme Inhib Med Chem, 35, 2020
|
|
1A7Z
 
 | | CRYSTAL STRUCTURE OF ACTINOMYCIN Z3 | | Descriptor: | ACTINOMYCIN Z3, BENZENE | | Authors: | Schafer, M. | | Deposit date: | 1998-03-19 | | Release date: | 1999-03-23 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Crystal Structures of Actinomycin D and Actinomycin Z3.
Angew.Chem.Int.Ed.Engl., 37, 1998
|
|
8AQG
 
 | |
5NMN
 
 | |
8AUU
 
 | |
8AE9
 
 | | Human Aldose Reductase in Complex with a Hydroxyphenyl-Thiophen-Acid Inhibitor (Schl32357) | | Descriptor: | 5-(5-(3-hydroxyphenyl)thiophen-2-yl)pentanoic acid, Aldose reductase, DIMETHYL SULFOXIDE, ... | | Authors: | Hubert, L.-S, Heine, A, Klebe, G, Reul, S. | | Deposit date: | 2022-07-12 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Human Aldose Reductase in Complex with a Hydroxyphenyl-Thiophen-Acid Inhibitor (Schl32357)
To Be Published
|
|
7VI4
 
 | | Electron crystallographic structure of TIA-1 prion-like domain, A381T mutant | | Descriptor: | TIA-1 prion-like domain | | Authors: | Takaba, K, Maki-Yonekura, S, Sekiyama, N, Imamura, K, Kodama, T, Tochio, H, Yonekura, K. | | Deposit date: | 2021-09-24 | | Release date: | 2022-09-28 | | Last modified: | 2024-06-19 | | Method: | ELECTRON CRYSTALLOGRAPHY (0.95 Å) | | Cite: | ALS mutations in the TIA-1 prion-like domain trigger highly condensed pathogenic structures.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
