7PY2
 
 | | Structure of pathological TDP-43 filaments from ALS with FTLD | | Descriptor: | TAR DNA-binding protein 43 | | Authors: | Arseni, D, Hasegawa, H, Murzin, A.G, Kametani, F, Arai, M, Yoshida, M, Falcon, B. | | Deposit date: | 2021-10-08 | | Release date: | 2021-12-15 | | Last modified: | 2022-01-19 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structure of pathological TDP-43 filaments from ALS with FTLD.
Nature, 601, 2022
|
|
8ANY
 
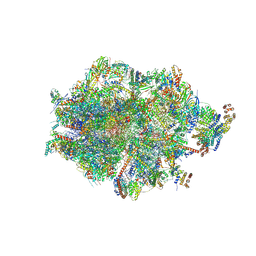 | | Human mitochondrial ribosome in complex with LRPPRC, SLIRP, A-site, P-site, E-site tRNAs and mRNA | | Descriptor: | 1,4-DIAMINOBUTANE, 12S mitochondrial rRNA, 16S mitochondrial rRNA, ... | | Authors: | Singh, V, Itoh, Y, Amunts, A. | | Deposit date: | 2022-08-06 | | Release date: | 2023-08-16 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Structural basis of LRPPRC-SLIRP-dependent translation by the
mitoribosome
To Be Published
|
|
7PU5
 
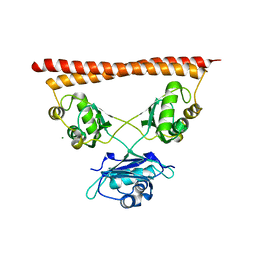 | | Structure of SFPQ-NONO complex | | Descriptor: | MAGNESIUM ION, Non-POU domain-containing octamer-binding protein, Splicing factor, ... | | Authors: | Fribourg, S. | | Deposit date: | 2021-09-28 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.999 Å) | | Cite: | Crystal structure of SFPQ-NONO heterodimer.
Biochimie, 198, 2022
|
|
7Q3L
 
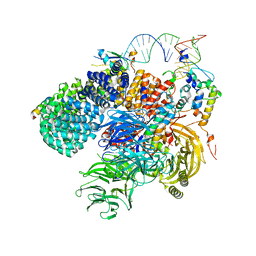 | | Human 17S U2 snRNP 5' domain | | Descriptor: | HIV Tat-specific factor 1, PHD finger-like domain-containing protein 5A, Probable ATP-dependent RNA helicase DDX46, ... | | Authors: | Tholen, J, Galej, W.P. | | Deposit date: | 2021-10-28 | | Release date: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Structural basis of branch site recognition by the human spliceosome.
Science, 375, 2022
|
|
7Q4O
 
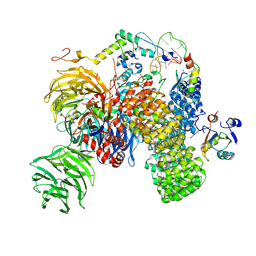 | | Substrate-bound A-like U2 snRNP | | Descriptor: | BPS oligo, PHD finger-like domain-containing protein 5A, Splicing factor 3A subunit 2, ... | | Authors: | Tholen, J, Galej, W.P. | | Deposit date: | 2021-11-01 | | Release date: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Structural basis of branch site recognition by the human spliceosome.
Science, 375, 2022
|
|
8BY6
 
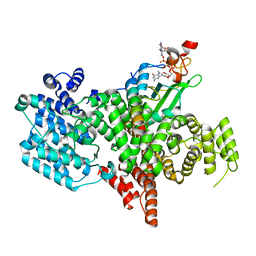 | | Structure of the human nuclear cap-binding complex bound to NCBP3(560-620) and cap-analogue m7GpppG | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-GUANOSINE, Nuclear cap-binding protein subunit 1, Nuclear cap-binding protein subunit 2, ... | | Authors: | Dubiez, E, Pellegrini, E, Foucher, A.E, Cusack, S, Kadlec, J. | | Deposit date: | 2022-12-12 | | Release date: | 2024-01-24 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Structural basis for competitive binding of productive and degradative co-transcriptional effectors to the nuclear cap-binding complex.
Cell Rep, 43, 2024
|
|
7YCX
 
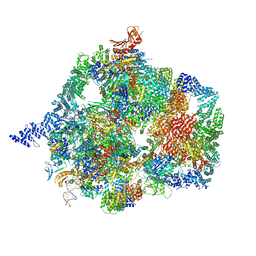 | | The structure of INTAC-PEC complex | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, DNA-directed RNA polymerase II subunit RPB1,DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Zheng, H, Jin, Q, Wang, X, Qi, Y, Liu, W, Ren, Y, Zhao, D, Chen, F.X, Cheng, J, Chen, X, Xu, Y. | | Deposit date: | 2022-07-02 | | Release date: | 2023-03-15 | | Last modified: | 2023-09-27 | | Method: | ELECTRON MICROSCOPY (4.18 Å) | | Cite: | Structural basis of INTAC-regulated transcription.
Protein Cell, 14, 2023
|
|
2VOP
 
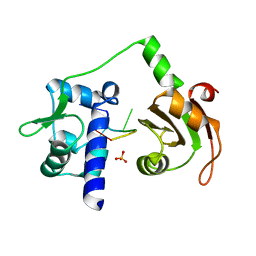 | | Crystal structure of N-terminal domains of Human La protein complexed with RNA oligomer AUUUU | | Descriptor: | 5'-R(*AP*UP*UP*UP*UP)-3', LUPUS LA PROTEIN, SULFATE ION | | Authors: | Kotik-Kogan, O, Valentine, E.R, Sanfelice, D, Conte, M.R, Curry, S. | | Deposit date: | 2008-02-19 | | Release date: | 2008-05-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Analysis Reveals Conformational Plasticity in the Recognition of RNA 3' Ends by the Human La Protein.
Structure, 16, 2008
|
|
2VOD
 
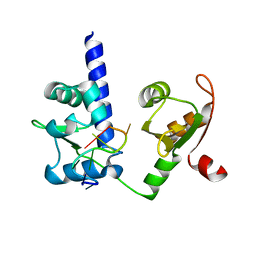 | | Crystal structure of N-terminal domains of Human La protein complexed with RNA oligomer AUAUUUU | | Descriptor: | 5'-R(*AP*UP*AP*UP*UP*UP*UP)-3', LUPUS LA PROTEIN | | Authors: | Kotik-Kogan, O, Valentine, E.R, Sanfelice, D, Conte, M.R, Curry, S. | | Deposit date: | 2008-02-15 | | Release date: | 2008-05-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Analysis Reveals Conformational Plasticity in the Recognition of RNA 3' Ends by the Human La Protein.
Structure, 16, 2008
|
|
2X1G
 
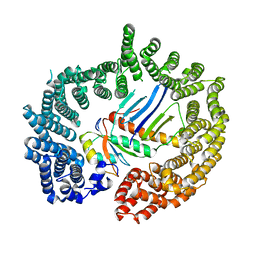 | | Crystal structure of Importin13 - Mago-Y14 complex | | Descriptor: | CADMUS, PROTEIN MAGO NASHI, RNA-BINDING PROTEIN 8A | | Authors: | Bono, F, Cook, A.G, Gruenwald, M, Ebert, J, Conti, E. | | Deposit date: | 2009-12-23 | | Release date: | 2010-02-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Nuclear Import Mechanism of the Ejc Component Mago- Y14 Revealed by Structural Studies of Importin 13.
Mol.Cell, 37, 2010
|
|
2XB2
 
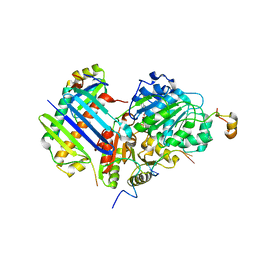 | | Crystal structure of the core Mago-Y14-eIF4AIII-Barentsz-UPF3b assembly shows how the EJC is bridged to the NMD machinery | | Descriptor: | EUKARYOTIC INITIATION FACTOR 4A-III, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Buchwald, G, Ebert, J, Basquin, C, Sauliere, J, Jayachandran, U, Bono, F, Le Hir, H, Conti, E. | | Deposit date: | 2010-04-03 | | Release date: | 2010-05-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Insights Into the Recruitment of the Nmd Machinery from the Crystal Structure of a Core Ejc-Upf3B Complex.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
7R6Q
 
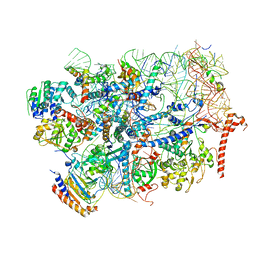 | | State E2 nucleolar 60S ribosome biogenesis intermediate - Foot region model | | Descriptor: | 25S rRNA, 5.8S rRNA, 60S ribosomal protein L13-A, ... | | Authors: | Cruz, V.E, Sekulski, K, Peddada, N, Erzberger, J.P. | | Deposit date: | 2021-06-23 | | Release date: | 2022-11-09 | | Last modified: | 2022-12-28 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Sequence-specific remodeling of a topologically complex RNP substrate by Spb4.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7R7A
 
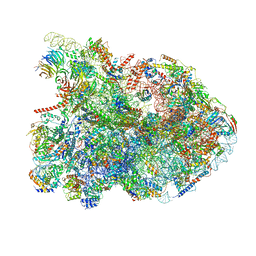 | | State E1 nucleolar 60S ribosome biogenesis intermediate - Composite model | | Descriptor: | 25S rRNA, 25S rRNA (cytosine(2870)-C(5))-methyltransferase, 27S pre-rRNA (guanosine(2922)-2'-O)-methyltransferase, ... | | Authors: | Cruz, V.E, Sekulski, K, Peddada, N, Erzberger, J.P. | | Deposit date: | 2021-06-24 | | Release date: | 2022-11-09 | | Last modified: | 2022-12-28 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Sequence-specific remodeling of a topologically complex RNP substrate by Spb4.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7SLP
 
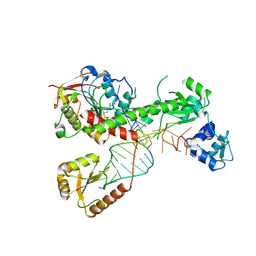 | | Cryo-EM structure of 7SK core RNP with linear RNA | | Descriptor: | 7SK snRNA methylphosphate capping enzyme, La-related protein 7, Linear 7SK RNA, ... | | Authors: | Yang, Y, Liu, S, Zhou, Z.H, Feigon, J. | | Deposit date: | 2021-10-24 | | Release date: | 2022-03-30 | | Last modified: | 2022-05-18 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis of RNA conformational switching in the transcriptional regulator 7SK RNP.
Mol.Cell, 82, 2022
|
|
7SLQ
 
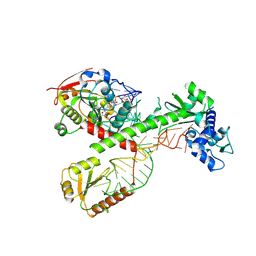 | | Cryo-EM structure of 7SK core RNP with circular RNA | | Descriptor: | 7SK snRNA methylphosphate capping enzyme, La-related protein 7, Minimal circular 7SK RNA, ... | | Authors: | Yang, Y, Liu, S, Zhou, Z.H, Feigon, J. | | Deposit date: | 2021-10-24 | | Release date: | 2022-03-30 | | Last modified: | 2022-05-18 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of RNA conformational switching in the transcriptional regulator 7SK RNP.
Mol.Cell, 82, 2022
|
|
7S7B
 
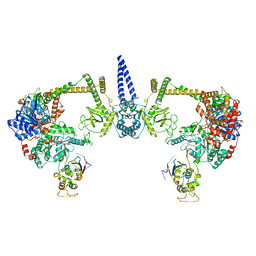 | |
7S7C
 
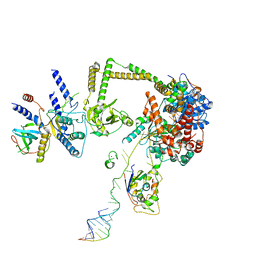 | |
7SP0
 
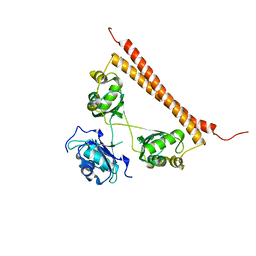 | | Crystal structure of human SFPQ L534I mutant in complex with zinc | | Descriptor: | Splicing factor, proline- and glutamine-rich, ZINC ION | | Authors: | Lee, M. | | Deposit date: | 2021-11-01 | | Release date: | 2022-10-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Familial ALS-associated SFPQ variants promote the formation of SFPQ cytoplasmic aggregates in primary neurons.
Open Biology, 12, 2022
|
|
7A5P
 
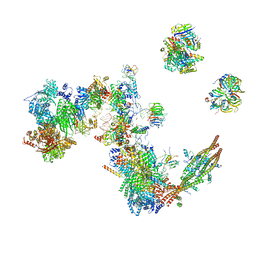 | | Human C Complex Spliceosome - Medium-resolution PERIPHERY | | Descriptor: | Cell division cycle 5-like protein, Crooked neck-like protein 1, Eukaryotic initiation factor 4A-III, ... | | Authors: | Bertram, K, Kastner, B. | | Deposit date: | 2020-08-21 | | Release date: | 2020-10-14 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | Structural Insights into the Roles of Metazoan-Specific Splicing Factors in the Human Step 1 Spliceosome.
Mol.Cell, 80, 2020
|
|
6ZON
 
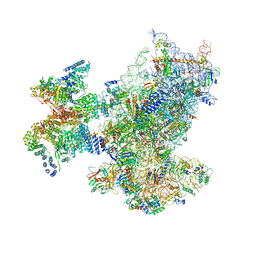 | | SARS-CoV-2 Nsp1 bound to a human 43S preinitiation ribosome complex - state 1 | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Thoms, M, Buschauer, R, Ameismeier, M, Denk, T, Kratzat, H, Mackens-Kiani, T, Cheng, J, Berninghausen, O, Becker, T, Beckmann, R. | | Deposit date: | 2020-07-07 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2.
Science, 369, 2020
|
|
7ABH
 
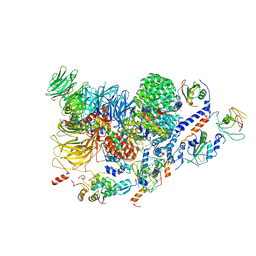 | | Human pre-Bact-2 spliceosome (SF3b/U2 snRNP portion) | | Descriptor: | Cell division cycle 5-like protein, DNA/RNA-binding protein KIN17, MINX M3 pre-mRNA, ... | | Authors: | Townsend, C, Kastner, B, Leelaram, M.N, Bertram, K, Stark, H, Luehrmann, R. | | Deposit date: | 2020-09-07 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Mechanism of protein-guided folding of the active site U2/U6 RNA during spliceosome activation.
Science, 370, 2020
|
|
7B0Y
 
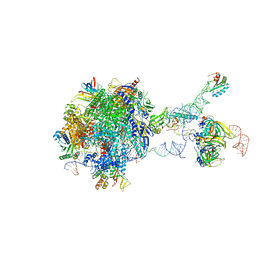 | | Structure of a transcribing RNA polymerase II-U1 snRNP complex | | Descriptor: | 145-nt RNA, DNA-directed RNA polymerase II subunit D, DNA-directed RNA polymerase II subunit E, ... | | Authors: | Zhang, S, Aibara, S, Vos, S.M, Agafonov, D.E, Luehrmann, R, Cramer, P. | | Deposit date: | 2020-11-23 | | Release date: | 2021-01-13 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure of a transcribing RNA polymerase II-U1 snRNP complex.
Science, 371, 2021
|
|
7C06
 
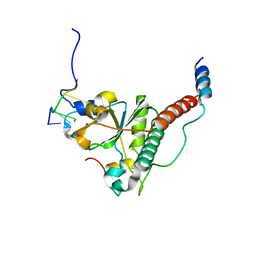 | | Crystal structure of yeast U2AF1 complex bound to 3' splice site RNA, 5'-UAGGU. | | Descriptor: | RNA (5'-R(*U*UP*AP*GP*GP*U)-3'), Splicing factor U2AF 23 kDa subunit, Splicing factor U2AF 59 kDa subunit, ... | | Authors: | Yoshida, H, Park, S.Y, Urano, T, Obayashi, E. | | Deposit date: | 2020-04-30 | | Release date: | 2020-09-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Elucidation of the aberrant 3' splice site selection by cancer-associated mutations on the U2AF1.
Nat Commun, 11, 2020
|
|
7C08
 
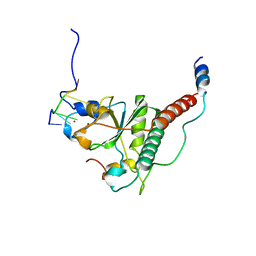 | | Crystal structure of S34Y mutant of yeast U2AF1 complex bound to 3' splice site RNA, 5'-UAGGU. | | Descriptor: | RNA (5'-R(*U*UP*AP*GP*GP*U)-3'), Splicing factor U2AF 23 kDa subunit, Splicing factor U2AF 59 kDa subunit, ... | | Authors: | Yoshida, H, Park, S.Y, Urano, T, Obayashi, E. | | Deposit date: | 2020-04-30 | | Release date: | 2020-09-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Elucidation of the aberrant 3' splice site selection by cancer-associated mutations on the U2AF1.
Nat Commun, 11, 2020
|
|
7BTB
 
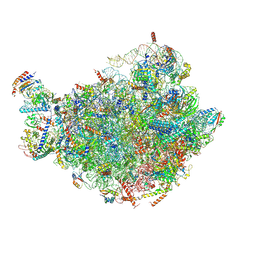 | |
