7SXH
 
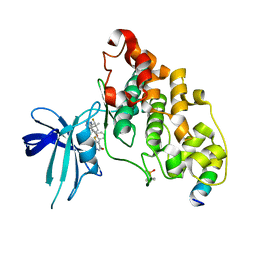 | | BIO-8546 bound GSK3beta-axin complex | | Descriptor: | (4S,5R,8R)-4-ethyl-8-fluoro-4-[3-(3-fluoro-5-methoxypyridin-4-yl)phenyl]-7,7-dimethyl-4,5,6,7,8,9-hexahydro-2H-pyrazolo[3,4-b]quinolin-5-ol, Glycogen synthase kinase-3 beta, axin peptide | | Authors: | Chodaparambil, J.V. | | Deposit date: | 2021-11-23 | | Release date: | 2023-06-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Elucidation of the GSK3 alpha Structure Informs the Design of Novel, Paralog-Selective Inhibitors.
Acs Chem Neurosci, 14, 2023
|
|
8SV9
 
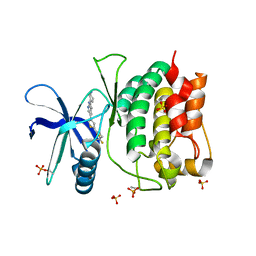 | | Crystal structure of ULK1 kinase domain with inhibitor MR-2088 | | Descriptor: | (4P)-4-[(2P)-2-(1,2,5,6-tetrahydropyridin-3-yl)-1H-pyrrolo[2,3-b]pyridin-5-yl]-N-(2,2,2-trifluoroethyl)thiophene-2-carboxamide, 1,2-ETHANEDIOL, SULFATE ION, ... | | Authors: | Schonbrunn, E, Sun, L. | | Deposit date: | 2023-05-15 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Development of potent and selective ULK1/2 inhibitors based on 7-azaindole scaffold with favorable in vivo properties.
Eur.J.Med.Chem., 266, 2024
|
|
7U36
 
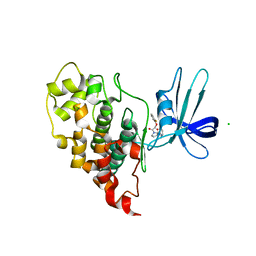 | | Crystal structure of human GSK3B in complex with ARN1484 | | Descriptor: | (3S)-1-[(2-fluorophenoxy)acetyl]-N-(pyridin-2-yl)pyrrolidine-3-carboxamide, CHLORIDE ION, Glycogen synthase kinase-3 beta | | Authors: | Tripathi, S.K, Balboni, B, Giabbai, B, Storici, P, Girotto, S, Cavalli, A. | | Deposit date: | 2022-02-25 | | Release date: | 2022-04-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Identification of Novel GSK-3 beta Hits Using Competitive Biophysical Assays.
Int J Mol Sci, 23, 2022
|
|
7U33
 
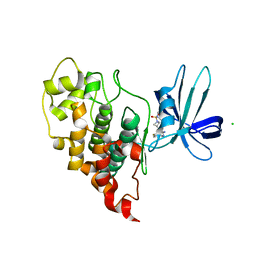 | | Crystal structure of human GSK3B in complex with ARN9133 | | Descriptor: | 3-[2-amino-5-(4-fluorophenyl)pyrimidin-4-yl]-N,N-dimethylazetidine-1-sulfonamide, CHLORIDE ION, Glycogen synthase kinase-3 beta | | Authors: | Tripathi, S.K, Balboni, B, Giabbai, B, Storici, P, Girotto, S, Cavalli, A. | | Deposit date: | 2022-02-25 | | Release date: | 2022-04-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Identification of Novel GSK-3 beta Hits Using Competitive Biophysical Assays.
Int J Mol Sci, 23, 2022
|
|
7U2Z
 
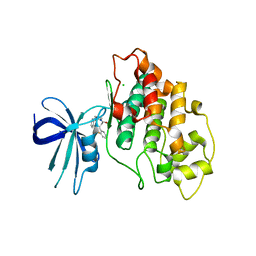 | | Crystal structure of human GSK3B in complex with G12 | | Descriptor: | (3R)-1-[3-(2-fluorophenyl)propanoyl]-N-(pyridin-2-yl)pyrrolidine-3-carboxamide, CHLORIDE ION, Glycogen synthase kinase-3 beta | | Authors: | Tripathi, S.K, Balboni, B, Giabbai, B, Storici, P, Girotto, S, Cavalli, A. | | Deposit date: | 2022-02-25 | | Release date: | 2022-04-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Identification of Novel GSK-3 beta Hits Using Competitive Biophysical Assays.
Int J Mol Sci, 23, 2022
|
|
8T2H
 
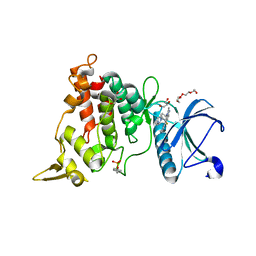 | | DYRK1A complex with DYR530 | | Descriptor: | (4P)-4-{(3M)-3-[3-fluoro-4-(4-methylpiperazin-1-yl)phenyl]-2-methyl-3H-imidazo[4,5-b]pyridin-5-yl}pyridin-2-amine, Dual specificity tyrosine-phosphorylation-regulated kinase 1A, GLYCEROL, ... | | Authors: | Montfort, W.R, Basantes, L.E. | | Deposit date: | 2023-06-06 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery of DYR684, a Potent, Selective, Metabolically Stable, DYRK1A/B PROTAC utilizing a Novel Cereblon Molecular Glue
To Be Published
|
|
8U2O
 
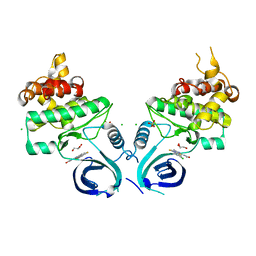 | |
8TUC
 
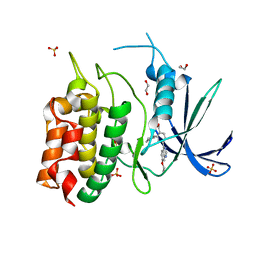 | | Unphosphorylated CaMKK2 in complex with CC-8977 | | Descriptor: | (4M)-2-cyclopentyl-4-(7-ethoxyquinazolin-4-yl)benzoic acid, 1,2-ETHANEDIOL, Calcium/calmodulin-dependent protein kinase kinase 2, ... | | Authors: | Bernard, S.M, Shanmugasundaram, V, D'Agostino, L. | | Deposit date: | 2023-08-16 | | Release date: | 2023-12-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Identification of Small Molecule Inhibitors and Ligand Directed Degraders of Calcium/Calmodulin Dependent Protein Kinase Kinase 1 and 2 (CaMKK1/2).
J.Med.Chem., 66, 2023
|
|
2NPQ
 
 | |
8U8J
 
 | |
7UP4
 
 | | Crystal structure of C-terminal Domain of MSK1 in complex with covalently bound pyrrolopyrimidine compound 20 (co-crystal) | | Descriptor: | (5M)-5-(2,5-dichloropyrimidin-4-yl)-5H-pyrrolo[3,2-d]pyrimidine, Ribosomal protein S6 kinase alpha-5 | | Authors: | Yano, J.K, Abendroth, J, Hall, A. | | Deposit date: | 2022-04-14 | | Release date: | 2022-07-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Discovery and Characterization of a Novel Series of Chloropyrimidines as Covalent Inhibitors of the Kinase MSK1.
Acs Med.Chem.Lett., 13, 2022
|
|
7UP6
 
 | | Crystal structure of C-terminal domain of MSK1 in complex with in covalently bound literature RSK2 inhibitor pyrrolopyrimidine cyanoacrylamide compound 25 (co-crystal) | | Descriptor: | (E)-3-(3-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)phenyl)-2-cyanoacrylamide bound form, OXAMIC ACID, Ribosomal protein S6 kinase alpha-5 | | Authors: | Yano, J.K, Abendroth, J, Hall, A. | | Deposit date: | 2022-04-14 | | Release date: | 2022-08-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery and Characterization of a Novel Series of Chloropyrimidines as Covalent Inhibitors of the Kinase MSK1.
Acs Med.Chem.Lett., 13, 2022
|
|
7UP8
 
 | |
7UP5
 
 | | Crystal structure of C-terminal Domain of MSK1 in complex with covalently bound pyrrolopyrimidine compound 23 (co-crystal) | | Descriptor: | (2M)-6-chloro-2-(5H-pyrrolo[3,2-d]pyrimidin-5-yl)pyridine-3-carbonitrile, IODIDE ION, Ribosomal protein S6 kinase alpha-5 | | Authors: | Yano, J.K, Edwards, T.E, Hall, A. | | Deposit date: | 2022-04-14 | | Release date: | 2022-07-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery and Characterization of a Novel Series of Chloropyrimidines as Covalent Inhibitors of the Kinase MSK1.
Acs Med.Chem.Lett., 13, 2022
|
|
8U8K
 
 | |
7UPM
 
 | |
2O5K
 
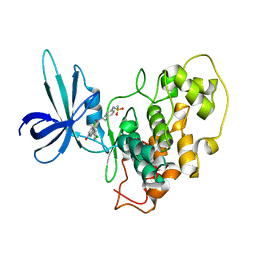 | | Crystal Structure of GSK3beta in complex with a benzoimidazol inhibitor | | Descriptor: | 2-(2,4-DICHLORO-PHENYL)-7-HYDROXY-1H-BENZOIMIDAZOLE-4-CARBOXYLIC ACID [2-(4-METHANESULFONYLAMINO-PHENYL)-ETHYL]-AMIDE, Glycogen synthase kinase-3 beta | | Authors: | Shin, D, Lee, S.C, Heo, Y.S, Cho, Y.S, Kim, Y.E, Hyun, Y.L, Cho, J.M, Lee, Y.S, Ro, S. | | Deposit date: | 2006-12-06 | | Release date: | 2007-10-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Design and synthesis of 7-hydroxy-1H-benzoimidazole derivatives as novel inhibitors of glycogen synthase kinase-3beta
Bioorg.Med.Chem.Lett., 17, 2007
|
|
8UDC
 
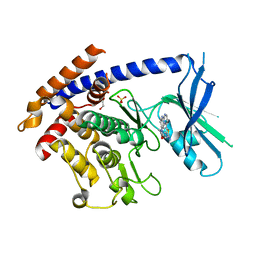 | | Crystal structure of TcPINK1 in complex with CYC116 | | Descriptor: | (4P)-4-(2-amino-4-methyl-1,3-thiazol-5-yl)-N-[4-(morpholin-4-yl)phenyl]pyrimidin-2-amine, DI(HYDROXYETHYL)ETHER, SULFATE ION, ... | | Authors: | Veyron, S, Rasool, S, Trempe, J.F. | | Deposit date: | 2023-09-28 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Characterization of a small-molecule inhibitor of PINK1, a precursor tool compound for the study of Parkinson's disease
To Be Published
|
|
8UCT
 
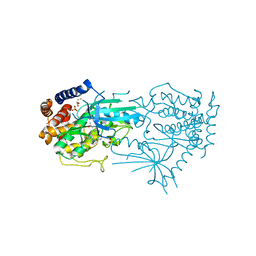 | | Crystal structure of TcPINK1 in complex with PRT | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 2-{[(1R,2S)-2-aminocyclohexyl]amino}-4-{[3-(2H-1,2,3-triazol-2-yl)phenyl]amino}pyrimidine-5-carboxamide, SULFATE ION, ... | | Authors: | Veyron, S, Rasool, S, Trempe, J.F. | | Deposit date: | 2023-09-27 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Characterization of a new family of PINK1 inhibitors
To Be Published
|
|
7UP7
 
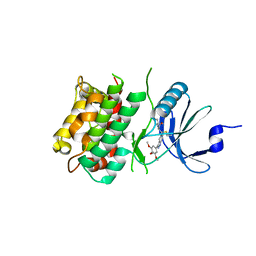 | | Crystal structure of C-terminal Domain of MSK1 in complex with covalently bound with literature RSK2 inhibitor indazole cyanoacrylamide compound 26 (soak) | | Descriptor: | (2S)-2-cyano-N-(1-hydroxy-2-methylpropan-2-yl)-3-[3-(3,4,5-trimethoxyphenyl)-1H-indazol-5-yl]propanamide, Ribosomal protein S6 kinase alpha-5 | | Authors: | Yano, J.K, Abendroth, J, Hall, A. | | Deposit date: | 2022-04-14 | | Release date: | 2022-07-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery and Characterization of a Novel Series of Chloropyrimidines as Covalent Inhibitors of the Kinase MSK1.
Acs Med.Chem.Lett., 13, 2022
|
|
7UKZ
 
 | |
2OKR
 
 | | Crystal Structure of the P38a-MAPKAP kinase 2 Heterodimer | | Descriptor: | MAP kinase-activated protein kinase 2, Mitogen-activated protein kinase 14 | | Authors: | Ter Haar, E. | | Deposit date: | 2007-01-17 | | Release date: | 2007-02-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the P38alpha-MAPKAP kinase 2 heterodimer.
J.Biol.Chem., 282, 2007
|
|
2O3P
 
 | | Crystal structure of Pim1 with Quercetin | | Descriptor: | 3,5,7,3',4'-PENTAHYDROXYFLAVONE, IMIDAZOLE, Proto-oncogene serine/threonine-protein kinase Pim-1 | | Authors: | Holder, S, Zemskova, M, Zhang, C, Tabrizizad, M, Bremer, R, Neidigh, J.W, Lilly, M.B. | | Deposit date: | 2006-12-01 | | Release date: | 2007-02-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Characterization of a potent and selective small-molecule inhibitor of the PIM1 kinase.
Mol.Cancer Ther., 6, 2007
|
|
2O65
 
 | | Crystal structure of Pim1 with Pentahydroxyflavone | | Descriptor: | 5,7-DIHYDROXY-2-(3,4,5-TRIHYDROXYPHENYL)-4H-CHROMEN-4-ONE, IMIDAZOLE, Proto-oncogene serine/threonine-protein kinase Pim-1 | | Authors: | Holder, S, Zemskova, M, Zhang, C, Tabrizizad, M, Bremer, R, Neidigh, J.W, Lilly, M.B. | | Deposit date: | 2006-12-06 | | Release date: | 2007-02-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Characterization of a potent and selective small-molecule inhibitor of the PIM1 kinase.
Mol.Cancer Ther., 6, 2007
|
|
2O64
 
 | | Crystal structure of Pim1 with Quercetagetin | | Descriptor: | 3,5,6,7-TETRAHYDROXY-2-(3,4-DIHYDROXYPHENYL)-4H-CHROMEN-4-ONE, IMIDAZOLE, Proto-oncogene serine/threonine-protein kinase Pim-1 | | Authors: | Holder, S, Zemskova, M, Zhang, C, Tabrizizad, M, Bremer, R, Neidigh, J.W, Lilly, M.B. | | Deposit date: | 2006-12-06 | | Release date: | 2007-02-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Characterization of a potent and selective small-molecule inhibitor of the PIM1 kinase.
Mol.Cancer Ther., 6, 2007
|
|
