8R04
 
 | | Structure of Staphylococcus aureus ClpP Bound to the Covalent Active Site Inhibitor Cystargolide A | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, Cystargolide A (bound) | | Authors: | Illigmann, A, Vielberg, M.-T, Lakemeyer, M, Wolf, F, Staudt, N, Dema, T, Stange, P, Liebhart, E, Kuttenlochner, W, Kulik, A, Malik, I, Grond, S, Sieber, S.A, Groll, M, Kaysser, L, Broetz-Oesterhelt, H. | | Deposit date: | 2023-10-30 | | Release date: | 2023-12-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of Staphylococcus aureus ClpP Bound to the Covalent Active-Site Inhibitor Cystargolide A.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
5NMS
 
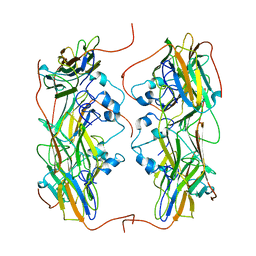 | | Hsp21 dodecamer, structural model based on cryo-EM and homology modelling | | Descriptor: | 25.3 kDa heat shock protein, chloroplastic | | Authors: | Rutsdottir, G, Harmark, J, Koeck, P.J.B, Hebert, H, Soderberg, C.A.G, Emanuelsson, C. | | Deposit date: | 2017-04-07 | | Release date: | 2017-05-03 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (10 Å) | | Cite: | Structural model of dodecameric heat-shock protein Hsp21: Flexible N-terminal arms interact with client proteins while C-terminal tails maintain the dodecamer and chaperone activity.
J. Biol. Chem., 292, 2017
|
|
4PE2
 
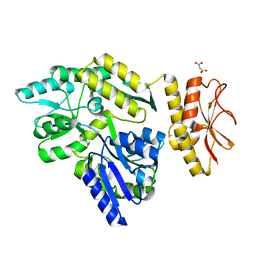 | | MBP PilA1 CD160 | | Descriptor: | MALONATE ION, Maltose ABC transporter periplasmic protein,Prepilin-type N-terminal cleavage/methylation domain protein, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Piepenbrink, K.H, Sundberg, E.J. | | Deposit date: | 2014-04-22 | | Release date: | 2015-01-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.724 Å) | | Cite: | Structural and Evolutionary Analyses Show Unique Stabilization Strategies in the Type IV Pili of Clostridium difficile.
Structure, 23, 2015
|
|
5NRN
 
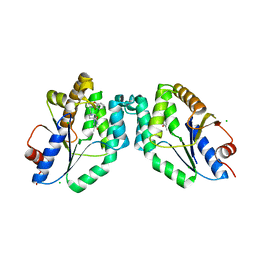 | | Mtb TMK crystal structure in complex with compound 3 | | Descriptor: | 5-methyl-1-[1-[(6-phenoxypyridin-2-yl)methyl]piperidin-4-yl]pyrimidine-2,4-dione, CHLORIDE ION, Thymidylate kinase | | Authors: | Merceron, R, Song, L, Munier-Lehmann, H, Van Calenbergh, S, Savvides, S. | | Deposit date: | 2017-04-25 | | Release date: | 2018-08-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mtb TMK crystal structure in complex with compound LS3080
To Be Published
|
|
4DR6
 
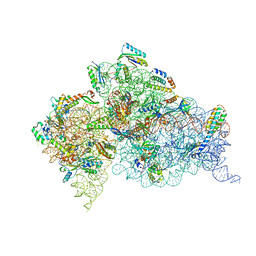 | | Crystal structure of the Thermus thermophilus (HB8) 30S ribosomal subunit with codon, near-cognate transfer RNA anticodon stem-loop mismatched at the first codon position and streptomycin bound | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Demirci, H, Murphy IV, F, Murphy, E, Gregory, S.T, Dahlberg, A.E, Jogl, G. | | Deposit date: | 2012-02-16 | | Release date: | 2012-11-14 | | Last modified: | 2013-01-30 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | A structural basis for streptomycin-induced misreading of the genetic code.
Nat Commun, 4, 2013
|
|
5NRT
 
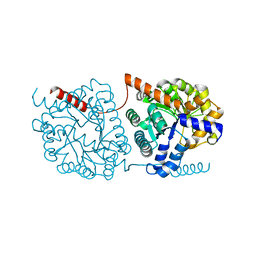 | | Cys-Gly dipeptidase GliJ in complex with Ca2+ | | Descriptor: | CALCIUM ION, Dipeptidase gliJ, MAGNESIUM ION | | Authors: | Groll, M, Huber, E.M. | | Deposit date: | 2017-04-25 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Gliotoxin Biosynthesis: Structure, Mechanism, and Metal Promiscuity of Carboxypeptidase GliJ.
ACS Chem. Biol., 12, 2017
|
|
5ZKL
 
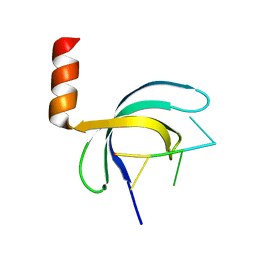 | | Crystal structure of Streptococcus pneumoniae SP_0782 (residues 7-79) in complex with single-stranded DNA dT12 | | Descriptor: | DNA (5'-D(*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), SP_0782 | | Authors: | Lu, G, Li, S, Zhu, J, Yang, Y, Gong, P. | | Deposit date: | 2018-03-24 | | Release date: | 2019-03-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.951 Å) | | Cite: | Structural insight into the length-dependent binding of ssDNA by SP_0782 from Streptococcus pneumoniae, reveals a divergence in the DNA-binding interface of PC4-like proteins.
Nucleic Acids Res., 48, 2020
|
|
5YVH
 
 | |
8OLL
 
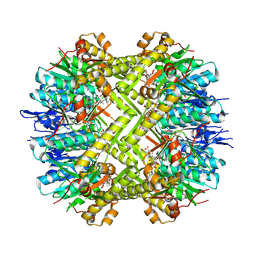 | | Staphylococcus aureus ClpP in complex with the natural product beta-lactone inhibitor Cystargolide A at 2.7 A resolution | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, Cystargolide A (bound) | | Authors: | Illigmann, A, Vielberg, M.-T, Lakemeyer, M, Wolf, F, Staudt, N, Dema, T, Stange, P, Malik, I, Grond, S, Sieber, S.A, Groll, M, Kaysser, L, Broetz-Oesterhelt, H. | | Deposit date: | 2023-03-30 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of Staphylococcus aureus ClpP Bound to the Covalent Active-Site Inhibitor Cystargolide A.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
5JZE
 
 | | Erve virus viral OTU domain protease in complex with mouse ISG15 | | Descriptor: | CITRATE ANION, RNA-dependent RNA polymerase, Ubiquitin-like protein ISG15, ... | | Authors: | Deaton, M.K, Dzimianski, J.V, Pegan, S.D. | | Deposit date: | 2016-05-16 | | Release date: | 2016-10-19 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Biochemical and Structural Insights into the Preference of Nairoviral DeISGylases for Interferon-Stimulated Gene Product 15 Originating from Certain Species.
J.Virol., 90, 2016
|
|
8OLR
 
 | | Structure of yeast 20S proteasome in complex with the natural product beta-lactone inhibitor Cystargolide A | | Descriptor: | CHLORIDE ION, Cystargolide A (bound), MAGNESIUM ION, ... | | Authors: | Illigmann, A, Vielberg, M.-T, Lakemeyer, M, Wolf, F, Staudt, N, Dema, T, Stange, P, Malik, I, Grond, S, Sieber, S.A, Groll, M, Kaysser, L, Broetz-Oesterhelt, H. | | Deposit date: | 2023-03-30 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of Staphylococcus aureus ClpP Bound to the Covalent Active-Site Inhibitor Cystargolide A.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
2H5K
 
 | | Crystal Structure of Complex Between the Domain-Swapped Dimeric Grb2 SH2 Domain and Shc-Derived Ligand, Ac-NH-pTyr-Val-Asn-NH2 | | Descriptor: | CACODYLATE ION, Growth factor receptor-bound protein 2, Shc-Derived Ligand | | Authors: | Benfield, A.P, Whiddon, B.B, Martin, S.F. | | Deposit date: | 2006-05-26 | | Release date: | 2006-08-15 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structural and energetic aspects of Grb2-SH2 domain-swapping.
Arch.Biochem.Biophys., 462, 2007
|
|
5LEZ
 
 | | Human 20S proteasome complex with Oprozomib in Mg-Acetate at 2.2 Angstrom | | Descriptor: | ACETATE ION, MAGNESIUM ION, PENTAETHYLENE GLYCOL, ... | | Authors: | Schrader, J, Henneberg, F, Mata, R, Tittmann, K, Schneider, T.R, Stark, H, Bourenkov, G, Chari, A. | | Deposit date: | 2016-06-30 | | Release date: | 2016-08-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | The inhibition mechanism of human 20S proteasomes enables next-generation inhibitor design.
Science, 353, 2016
|
|
8QWB
 
 | |
5LKM
 
 | | RadA bound to dTDP | | Descriptor: | DNA repair protein RadA, MAGNESIUM ION, THYMIDINE-5'-DIPHOSPHATE | | Authors: | Rapisarda, C, Remaut, H, Fornzes, R. | | Deposit date: | 2016-07-22 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Bacterial RadA is a DnaB-type helicase interacting with RecA to promote bidirectional D-loop extension.
Nat Commun, 8, 2017
|
|
2PKS
 
 | | Thrombin in complex with inhibitor | | Descriptor: | 4-({[4-(3-METHYLBENZOYL)PYRIDIN-2-YL]AMINO}METHYL)BENZENECARBOXIMIDAMIDE, Hirudin, SODIUM ION, ... | | Authors: | Xue, Y. | | Deposit date: | 2007-04-18 | | Release date: | 2008-04-22 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Design, synthesis and biological evaluation of thrombin inhibitors based on a pyridine scaffold.
Org.Biomol.Chem., 5, 2007
|
|
5WB8
 
 | | Crystal structure of the epidermal growth factor receptor extracellular region in complex with epigen | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Epidermal growth factor receptor, Epigen, ... | | Authors: | Bessman, N.J, Freed, D.M, Moore, J.O, Ferguson, K.M, Lemmon, M.A. | | Deposit date: | 2017-06-28 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | EGFR Ligands Differentially Stabilize Receptor Dimers to Specify Signaling Kinetics.
Cell, 171, 2017
|
|
5WEW
 
 | | Crystal structure of Klebsiella pneumoniae fosfomycin resistance protein (FosAKP) with inhibitor (ANY1) bound | | Descriptor: | 6,6'-(4-nitro-1H-pyrazole-3,5-diyl)bis(3-bromopyrazolo[1,5-a]pyrimidin-2(1H)-one), Fosfomycin resistance protein, MANGANESE (II) ION | | Authors: | Klontz, E.H, Sundberg, E.J. | | Deposit date: | 2017-07-10 | | Release date: | 2018-07-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.178 Å) | | Cite: | Small-Molecule Inhibitor of FosA Expands Fosfomycin Activity to Multidrug-Resistant Gram-Negative Pathogens.
Antimicrob. Agents Chemother., 63, 2019
|
|
5WK6
 
 | |
7X3D
 
 | | Cryo-EM structure of Coxsackievirus B1 mature virion in complex with nAb 9A3 (CVB1-M:9A3) | | Descriptor: | 9A3 heavy chain, 9A3 light chain, Capsid protein VP4, ... | | Authors: | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | Deposit date: | 2022-02-28 | | Release date: | 2022-09-28 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
7X40
 
 | | Cryo-EM structure of Coxsackievirus B1 mature virion in complex with nAb 8A10 (classified from CVB1 mature virion in complex with 8A10 and 2E6) | | Descriptor: | 8A10 heavy chain, 8A10 light chain, Capsid protein VP4, ... | | Authors: | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | Deposit date: | 2022-03-01 | | Release date: | 2022-09-28 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
7X46
 
 | | Cryo-EM structure of Coxsackievirus B1 A-particle in complex with nAb 2E6 (classified from CVB1 mature virion in complex with 8A10 and 2E6) | | Descriptor: | 2E6 heavy chain, 2E6 light chain, VP2, ... | | Authors: | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | Deposit date: | 2022-03-02 | | Release date: | 2022-09-28 | | Method: | ELECTRON MICROSCOPY (3.85 Å) | | Cite: | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
7X47
 
 | | Cryo-EM structure of Coxsackievirus B1 empty particle in complex with nAb 2E6 (classified from CVB1 mature virion in complex with 8A10 and 2E6) | | Descriptor: | 2E6 heavy chain, 2E6 light chain, Genome polyprotein, ... | | Authors: | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | Deposit date: | 2022-03-02 | | Release date: | 2022-09-28 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
7X4K
 
 | | Cryo-EM structure of Coxsackievirus B1 empty particle in complex with nAb 9A3 (classified from CVB1 mature virion in complex with 8A10 and 9A3) | | Descriptor: | 9A3 heavy chain, 9A3 light chain, Genome polyprotein, ... | | Authors: | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | Deposit date: | 2022-03-02 | | Release date: | 2022-09-28 | | Method: | ELECTRON MICROSCOPY (3.82 Å) | | Cite: | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
4DR7
 
 | | Crystal structure of the Thermus thermophilus (HB8) 30S ribosomal subunit with codon, crystallographically disordered near-cognate transfer RNA anticodon stem-loop mismatched at the second codon position, and streptomycin bound | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Demirci, H, Murphy IV, F, Murphy, E, Gregory, S.T, Dahlberg, A.E, Jogl, G. | | Deposit date: | 2012-02-16 | | Release date: | 2012-11-14 | | Last modified: | 2013-01-30 | | Method: | X-RAY DIFFRACTION (3.75 Å) | | Cite: | A structural basis for streptomycin-induced misreading of the genetic code.
Nat Commun, 4, 2013
|
|
