7XHQ
 
 | | Small-molecule Allosteric Regulation Mechanism of SHP2 | | Descriptor: | 2-[(3S,4R)-4-azanyl-3-methyl-2-oxa-8-azaspiro[4.5]decan-8-yl]-5-[2,3-bis(chloranyl)phenyl]-3-methyl-pyrrolo[2,1-f][1,2,4]triazin-4-one, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Luo, Y, Zhu, J, Yu, K, Liu, B. | | Deposit date: | 2022-04-09 | | Release date: | 2023-02-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of the SHP2 allosteric inhibitor 2-((3R,4R)-4-amino-3-methyl-2-oxa-8-azaspiro[4.5]decan-8-yl)-5-(2,3-dichlorophenyl)-3-methylpyrrolo[2,1-f][1,2,4] triazin-4(3H)-one.
J Enzyme Inhib Med Chem, 38, 2023
|
|
8CBH
 
 | | SHP2 in complex with a novel allosteric inhibitor | | Descriptor: | Tyrosine-protein phosphatase non-receptor type 11, [(1~{S},6~{R},7~{S})-3-[3-[2,3-bis(chloranyl)phenyl]-2~{H}-pyrazolo[3,4-b]pyrazin-6-yl]-7-(4-methyl-1,3-thiazol-2-yl)-3-azabicyclo[4.1.0]heptan-7-yl]methanamine | | Authors: | di Fabio, R, Petrocchi, A. | | Deposit date: | 2023-01-25 | | Release date: | 2023-04-26 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Discovery of a Novel Series of Imidazopyrazine Derivatives as Potent SHP2 Allosteric Inhibitors.
Acs Med.Chem.Lett., 14, 2023
|
|
7ZLO
 
 | | Crystal structure of SOCS2:ElonginB:ElonginC in complex with compound 12 | | Descriptor: | Elongin-B, Elongin-C, Suppressor of cytokine signaling 2, ... | | Authors: | Ramachandran, S, Ciulli, A, Makukhin, N. | | Deposit date: | 2022-04-15 | | Release date: | 2023-04-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structure-based design of a phosphotyrosine-masked covalent ligand targeting the E3 ligase SOCS2.
Nat Commun, 14, 2023
|
|
7ZLP
 
 | | Crystal structure of SOCS2:ElonginB:ElonginC in complex with compound 9 | | Descriptor: | Elongin-B, Elongin-C, PHOSPHATE ION, ... | | Authors: | Ramachandran, S, Ciulli, A, Makukhin, N. | | Deposit date: | 2022-04-15 | | Release date: | 2023-04-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structure-based design of a phosphotyrosine-masked covalent ligand targeting the E3 ligase SOCS2.
Nat Commun, 14, 2023
|
|
7ZLN
 
 | | Crystal structure of SOCS2:ElonginB:ElonginC in complex with compound 11 | | Descriptor: | Elongin-B, Elongin-C, Suppressor of cytokine signaling 2, ... | | Authors: | Ramachandran, S, Ciulli, A, Makukhin, N. | | Deposit date: | 2022-04-15 | | Release date: | 2023-04-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-based design of a phosphotyrosine-masked covalent ligand targeting the E3 ligase SOCS2.
Nat Commun, 14, 2023
|
|
7ZLS
 
 | | co-crystal structure of SOCS2:ElonginB:ElonginC in complex with compound 13 | | Descriptor: | 1,2-ETHANEDIOL, Elongin-B, Elongin-C, ... | | Authors: | Ramachandran, S, Ciulli, A, Makukhin, N. | | Deposit date: | 2022-04-15 | | Release date: | 2023-04-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structure-based design of a phosphotyrosine-masked covalent ligand targeting the E3 ligase SOCS2.
Nat Commun, 14, 2023
|
|
7ZLR
 
 | | Crystal structure of SOCS2:ElonginB:ElonginC in complex with compound 13 | | Descriptor: | Elongin-B, Elongin-C, Suppressor of cytokine signaling 2, ... | | Authors: | Ramachandran, S, Ciulli, A, Makukhin, N. | | Deposit date: | 2022-04-15 | | Release date: | 2023-04-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structure-based design of a phosphotyrosine-masked covalent ligand targeting the E3 ligase SOCS2.
Nat Commun, 14, 2023
|
|
7ZLM
 
 | | Crystal structure of SOCS2:ElonginB:ElonginC in complex with compound MN551 | | Descriptor: | Elongin-B, Elongin-C, Suppressor of cytokine signaling 2, ... | | Authors: | Ramachandran, S, Ciulli, A, Makukhin, N. | | Deposit date: | 2022-04-15 | | Release date: | 2023-04-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structure-based design of a phosphotyrosine-masked covalent ligand targeting the E3 ligase SOCS2.
Nat Commun, 14, 2023
|
|
8ATK
 
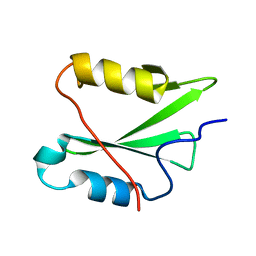 | | The SH2 domain of mouse SH2B1 | | Descriptor: | SH2B adapter protein 1 | | Authors: | Fowler, N.J, Williamson, M.P, Albalwi, M.F. | | Deposit date: | 2022-08-23 | | Release date: | 2023-05-24 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Improved methodology for protein NMR structure calculation using hydrogen bond restraints and ANSURR validation: The SH2 domain of SH2B1.
Structure, 31, 2023
|
|
8CZ9
 
 | |
8DGO
 
 | |
8SBJ
 
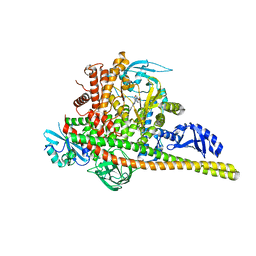 | | Co-structure Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform complexed with brain penetrant inhibitors | | Descriptor: | (2M)-7-[(3R)-3-methylmorpholin-4-yl]-5-[(3S)-3-methylmorpholin-4-yl]-2-(1H-pyrazol-3-yl)-3H-imidazo[4,5-b]pyridine, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Knapp, M.S, Elling, R.A, Tang, J. | | Deposit date: | 2023-04-03 | | Release date: | 2023-07-19 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Identification of Brain-Penetrant ATP-Competitive mTOR Inhibitors for CNS Syndromes.
J.Med.Chem., 66, 2023
|
|
8SBC
 
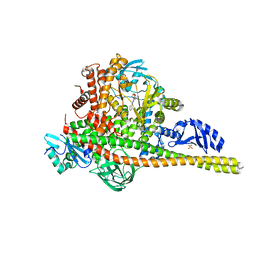 | | Co-structure of Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform and brain penetrant inhibitors | | Descriptor: | (2M)-7-[(3R)-3-methylmorpholin-4-yl]-5-[(3S)-3-methylmorpholin-4-yl]-2-(pyridin-2-yl)-1H-imidazo[4,5-b]pyridine, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform, ... | | Authors: | Knapp, M.S, Elling, R.A, Tang, J. | | Deposit date: | 2023-04-03 | | Release date: | 2023-07-19 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification of Brain-Penetrant ATP-Competitive mTOR Inhibitors for CNS Syndromes.
J.Med.Chem., 66, 2023
|
|
8GMB
 
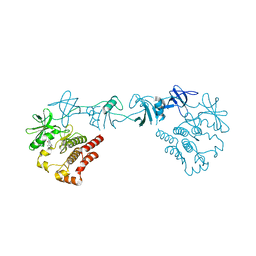 | | Crystal structure of the full-length Bruton's tyrosine kinase (PH-TH domain not visible) | | Descriptor: | 2-[3'-(hydroxymethyl)-1-methyl-5-({5-[(2S)-2-methyl-4-(oxetan-3-yl)piperazin-1-yl]pyridin-2-yl}amino)-6-oxo[1,6-dihydro[3,4'-bipyridine]]-2'-yl]-7,7-dimethyl-3,4,7,8-tetrahydro-2H-cyclopenta[4,5]pyrrolo[1,2-a]pyrazin-1(6H)-one, Tyrosine-protein kinase BTK | | Authors: | Lin, D.Y, Andreotti, A.H. | | Deposit date: | 2023-03-24 | | Release date: | 2023-08-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Conformational heterogeneity of the BTK PHTH domain drives multiple regulatory states.
Elife, 12, 2024
|
|
7YQE
 
 | |
8ILR
 
 | | Cryo-EM structure of PI3Kalpha in complex with compound 16 | | Descriptor: | N-[(2S)-1-(ethylamino)-1-oxidanylidene-3-[4-(2-quinoxalin-6-ylethynyl)phenyl]propan-2-yl]-2,3-dimethyl-quinoxaline-6-carboxamide, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Zhou, Q, Liu, X, Neri, D, Li, W, Favalli, N, Bassi, G, Yang, S, Yang, D, Vogt, P.K, Wang, M.-W. | | Deposit date: | 2023-03-04 | | Release date: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Structural insights into the interaction of three Y-shaped ligands with PI3K alpha.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8TGD
 
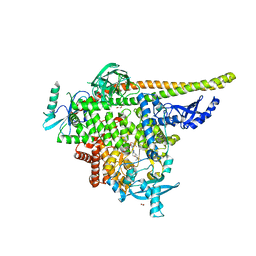 | | STX-478, a Mutant-Selective, Allosteric Inhibitor bound to H1047R PI3Kalpha | | Descriptor: | 1,2-ETHANEDIOL, N-(2-aminopyrimidin-5-yl)-N'-[(1R)-1-(5,7-difluoro-3-methyl-1-benzofuran-2-yl)-2,2,2-trifluoroethyl]urea, N~2~-{(4S,11aP)-2-[(4S)-4-(difluoromethyl)-2-oxo-1,3-oxazolidin-3-yl]-5,6-dihydroimidazo[1,2-d][1,4]benzoxazepin-9-yl}-L-alaninamide, ... | | Authors: | Hilbert, B, Brooijmans, N, Buckbinder, L, St.Jean Jr, D.J. | | Deposit date: | 2023-07-12 | | Release date: | 2023-09-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.928 Å) | | Cite: | STX-478, a Mutant-Selective, Allosteric PI3K alpha Inhibitor Spares Metabolic Dysfunction and Improves Therapeutic Response in PI3K alpha-Mutant Xenografts.
Cancer Discov, 13, 2023
|
|
8SSN
 
 | | Abl kinase in complex with SKI and asciminib | | Descriptor: | 6,7-dimethoxy-N-(4-phenoxyphenyl)quinazolin-4-amine, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Ludewig, H, Kim, C, Kern, D. | | Deposit date: | 2023-05-08 | | Release date: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | A biophysical framework for double-drugging kinases.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8TDU
 
 | | STX-478, a Mutant-Selective, Allosteric Inhibitor bound to PI3Kalpha | | Descriptor: | N-(2-aminopyrimidin-5-yl)-N'-[(1R)-1-(5,7-difluoro-3-methyl-1-benzofuran-2-yl)-2,2,2-trifluoroethyl]urea, N~2~-{(4S,11aP)-2-[(4S)-4-(difluoromethyl)-2-oxo-1,3-oxazolidin-3-yl]-5,6-dihydroimidazo[1,2-d][1,4]benzoxazepin-9-yl}-L-alaninamide, Phosphatidylinositol 3-kinase regulatory subunit alpha, ... | | Authors: | Hilbert, B.J, Brooijmans, N, Buckbinder, L, St.Jean Jr, D.J. | | Deposit date: | 2023-07-05 | | Release date: | 2023-09-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | STX-478, a Mutant-Selective, Allosteric PI3K alpha Inhibitor Spares Metabolic Dysfunction and Improves Therapeutic Response in PI3K alpha-Mutant Xenografts.
Cancer Discov, 13, 2023
|
|
8ILV
 
 | | Cryo-EM structure of PI3Kalpha in complex with compound 18 | | Descriptor: | N-[(2R)-1-(ethylamino)-1-oxidanylidene-3-[3-(2-quinoxalin-6-ylethynyl)phenyl]propan-2-yl]-2,3-dimethyl-quinoxaline-6-carboxamide, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Zhou, Q, Liu, X, Neri, D, Li, W, Favalli, N, Bassi, G, Yang, S, Yang, D, Vogt, P.K, Wang, M.-W. | | Deposit date: | 2023-03-04 | | Release date: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Structural insights into the interaction of three Y-shaped ligands with PI3K alpha.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8ILS
 
 | | Cryo-EM structure of PI3Kalpha in complex with compound 17 | | Descriptor: | N-[(2R)-1-(ethylamino)-1-oxidanylidene-3-[4-(2-quinoxalin-6-ylethynyl)phenyl]propan-2-yl]-2,3-dimethyl-quinoxaline-6-carboxamide, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Zhou, Q, Liu, X, Neri, D, Li, W, Favalli, N, Bassi, G, Yang, S, Yang, D, Vogt, P.K, Wang, M.-W. | | Deposit date: | 2023-03-04 | | Release date: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into the interaction of three Y-shaped ligands with PI3K alpha.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8GWW
 
 | | Small-molecule Allosteric Regulation Mechanism of SHP2 | | Descriptor: | 2-[4-(aminomethyl)-4-methyl-piperidin-1-yl]-5-[2,3-bis(chloranyl)phenyl]-3-methyl-pyrrolo[2,1-f][1,2,4]triazin-4-one, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Luo, Y, Zhu, J, Yu, K, Liu, B. | | Deposit date: | 2022-09-17 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Small-molecule Allosteric Regulation Mechanism of SHP2
To Be Published
|
|
8T8Q
 
 | | Identification of GDC-1971 (RLY-1971), a SHP2 inhibitor designed for the treatment of solid tumors | | Descriptor: | 1-[(3P)-3-(3-chloro-2-fluorophenyl)-1H-pyrazolo[3,4-b]pyrazin-6-yl]-4-methylpiperidin-4-amine, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Tang, Y, Nguyen, V, Wilbur, J.D. | | Deposit date: | 2023-06-23 | | Release date: | 2023-10-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Identification of GDC-1971 (RLY-1971), a SHP2 Inhibitor Designed for the Treatment of Solid Tumors.
J.Med.Chem., 66, 2023
|
|
8T7Q
 
 | | Identification of GDC-1971 (RLY-1971), a SHP2 inhibitor designed for the treatment of solid tumors | | Descriptor: | 1-{3-[(2-chlorophenyl)sulfanyl]-1H-pyrazolo[3,4-b]pyrazin-6-yl}-4-methylpiperidin-4-amine, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Tang, Y, Nguyen, V, Wilbur, J.D. | | Deposit date: | 2023-06-21 | | Release date: | 2023-10-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification of GDC-1971 (RLY-1971), a SHP2 Inhibitor Designed for the Treatment of Solid Tumors.
J.Med.Chem., 66, 2023
|
|
8T6D
 
 | | Identification of GDC-1971 (RLY-1971), a SHP2 inhibitor designed for the treatment of solid tumors | | Descriptor: | (3R)-1'-[3-(3,4-dihydro-1,5-naphthyridin-1(2H)-yl)-1H-pyrazolo[3,4-b]pyrazin-6-yl]-3H-spiro[[1]benzofuran-2,4'-piperidin]-3-amine, SULFATE ION, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Tang, Y, Nguyen, V, Wilbur, J.D. | | Deposit date: | 2023-06-15 | | Release date: | 2023-10-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Identification of GDC-1971 (RLY-1971), a SHP2 Inhibitor Designed for the Treatment of Solid Tumors.
J.Med.Chem., 66, 2023
|
|
