8VXD
 
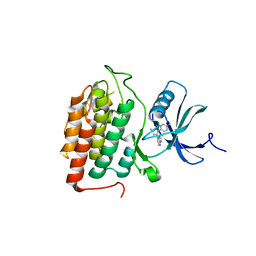 | |
8VXF
 
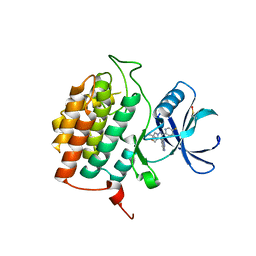 | | Structure of Casein kinase I isoform delta (CK1d) complexed with inhibitor 15 | | Descriptor: | (2P,3P,8S)-2-(5-fluoropyridin-2-yl)-6,6-dimethyl-3-(1H-pyrazolo[3,4-b]pyridin-4-yl)-6,7-dihydro-4H-pyrazolo[5,1-c][1,4]oxazine, Casein kinase I isoform delta | | Authors: | Thompson, A.A, Milligan, C.M, Sharma, S. | | Deposit date: | 2024-02-04 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structure-Based Optimization of Selective and Brain Penetrant CK1 delta Inhibitors for the Treatment of Circadian Disruptions.
Acs Med.Chem.Lett., 15, 2024
|
|
8VXE
 
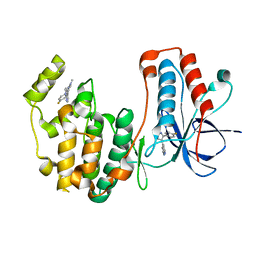 | | Structure of p38 alpha (Mitogen-activated protein kinase 14) complexed with inhibitor 6 | | Descriptor: | (4M)-4-[3-(4-fluorophenyl)-1-methyl-1H-pyrazol-4-yl]-1H-pyrrolo[2,3-b]pyridine, Mitogen-activated protein kinase 14 | | Authors: | Blaesse, M, Steinbacher, S, Shaffer, P.L, Sharma, S, Thompson, A.A. | | Deposit date: | 2024-02-04 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-Based Optimization of Selective and Brain Penetrant CK1 delta Inhibitors for the Treatment of Circadian Disruptions.
Acs Med.Chem.Lett., 15, 2024
|
|
8RW1
 
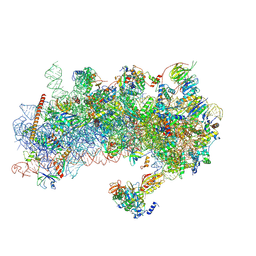 | | Structure of a yeast 48S-AUC preinitiation complex in closed conformation | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein L41-A, 40S ribosomal protein S12, ... | | Authors: | Villamayor-Belinchon, L, Sharma, P, Llacer, J.L, Hussain, T. | | Deposit date: | 2024-02-02 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Structural basis of AUC codon discrimination during translation initiation in yeast.
Nucleic Acids Res., 52, 2024
|
|
8Y6H
 
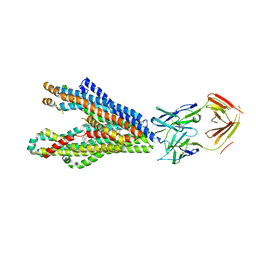 | | P-glycoprotein in complex with UIC2 Fab and triple elacridar molecules in LMNG detergent | | Descriptor: | ATP-dependent translocase ABCB1,mNeonGreen, UIC2 Fab heavy chain, UIC2 Fab light chain, ... | | Authors: | Hamaguchi-Suzuki, N, Adachi, N, Moriya, T, Kawasaki, M, Suzuki, K, Anzai, N, Senda, T, Murata, T. | | Deposit date: | 2024-02-02 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (2.49 Å) | | Cite: | Cryo-EM structure of P-glycoprotein bound to triple elacridar inhibitor molecules.
Biochem.Biophys.Res.Commun., 709, 2024
|
|
8Y6I
 
 | | P-glycoprotein in complex with UIC2 Fab and triple elacridar molecules in nanodisc | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, ATP-dependent translocase ABCB1,mNeonGreen, CHOLESTEROL, ... | | Authors: | Hamaguchi-Suzuki, N, Adachi, N, Moriya, T, Kawasaki, M, Suzuki, K, Anzai, N, Senda, T, Murata, T. | | Deposit date: | 2024-02-02 | | Release date: | 2024-04-17 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | Cryo-EM structure of P-glycoprotein bound to triple elacridar inhibitor molecules.
Biochem.Biophys.Res.Commun., 709, 2024
|
|
8Y6O
 
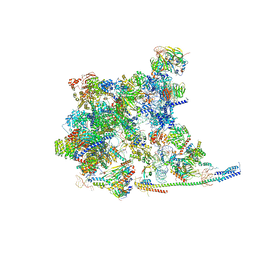 | | Cryo-EM Structure of the human minor pre-B complex (pre-precatalytic spliceosome) U11 and tri-snRNP part | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, Centrosomal AT-AC splicing factor, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Bai, R, Yuan, M, Zhang, P, Luo, T, Shi, Y, Wan, R. | | Deposit date: | 2024-02-02 | | Release date: | 2024-03-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Structural basis of U12-type intron engagement by the fully assembled human minor spliceosome.
Science, 383, 2024
|
|
8RVT
 
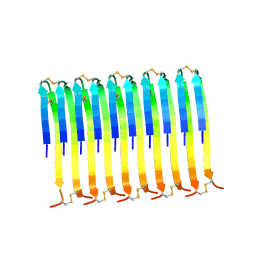 | |
8VX0
 
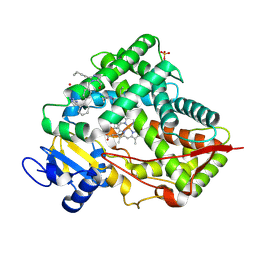 | | CRYSTAL STRUCTURE OF CYP2C9*14 IN COMPLEX WITH LOSARTAN | | Descriptor: | Cytochrome P450 2C9, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Shah, M.B. | | Deposit date: | 2024-02-02 | | Release date: | 2024-06-12 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structural and biophysical analysis of cytochrome P450 2C9*14 and *27 variants in complex with losartan.
J.Inorg.Biochem., 258, 2024
|
|
8Y66
 
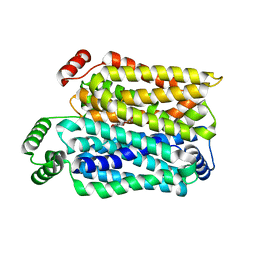 | | Cryo-EM structure of human urate transporter GLUT9 bound to inhibitor apigenin | | Descriptor: | 5,7-dihydroxy-2-(4-hydroxyphenyl)-4H-chromen-4-one, Solute carrier family 2, facilitated glucose transporter member 9 | | Authors: | Pan, X.J, Shen, Z.L, Xu, L, Huang, G.X.Y. | | Deposit date: | 2024-02-01 | | Release date: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Structural basis for urate recognition and apigenin inhibition of human GLUT9.
Nat Commun, 15, 2024
|
|
8Y65
 
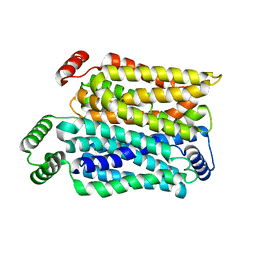 | | Cryo-EM structure of human urate transporter GLUT9 bound to substrate urate | | Descriptor: | Solute carrier family 2, facilitated glucose transporter member 9, URIC ACID | | Authors: | Pan, X.J, Shen, Z.L, Xu, L, Huang, G.X.Y. | | Deposit date: | 2024-02-01 | | Release date: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.51 Å) | | Cite: | Structural basis for urate recognition and apigenin inhibition of human GLUT9.
Nat Commun, 15, 2024
|
|
8RVE
 
 | | Vimentin intermediate filament | | Descriptor: | Vimentin | | Authors: | Eibauer, M, Medalia, O. | | Deposit date: | 2024-02-01 | | Release date: | 2024-04-10 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (7.2 Å) | | Cite: | Vimentin filaments integrate low-complexity domains in a complex helical structure.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8VVM
 
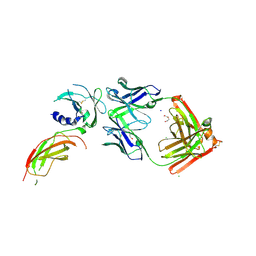 | | Structure of FabS1CE1-EPR1-1 in complex with the erythropoietin receptor | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Singer, A.U, Bruce, H.A, Pavlenco, A, Ploder, L, Luu, G, Blazer, L, Adams, J.J, Sidhu, S.S. | | Deposit date: | 2024-01-31 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Antigen-binding fragments with improved crystal lattice packing and enhanced conformational flexibility at the elbow region as crystallization chaperones.
Protein Sci., 33, 2024
|
|
8VVO
 
 | | Structure of FabS1CE2-EPR1-1 in complex with the erythropoietin receptor | | Descriptor: | CHLORIDE ION, Erythropoietin receptor, S1CE2 VARIANT OF FAB-EPR-1 heavy chain, ... | | Authors: | Singer, A.U, Bruce, H.A, Pavlenco, A, Ploder, L, Luu, G, Blazer, L, Adams, J.J, Sidhu, S.S. | | Deposit date: | 2024-01-31 | | Release date: | 2024-07-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Antigen-binding fragments with improved crystal lattice packing and enhanced conformational flexibility at the elbow region as crystallization chaperones.
Protein Sci., 33, 2024
|
|
8RUS
 
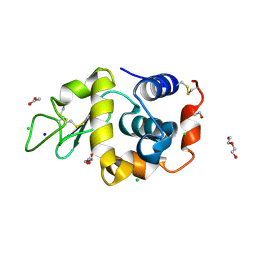 | | Hen egg-white lysozyme (HEWL) structure from EuXFEL FXE, multi-hit Droplet-on-Demand (DoD) injection, 9.3 keV photon energy, space group P432121 | | Descriptor: | ACETATE ION, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Perrett, S, van Thor, J.J. | | Deposit date: | 2024-01-31 | | Release date: | 2024-07-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Kilohertz droplet-on-demand serial femtosecond crystallography at the European XFEL station FXE.
Struct Dyn., 11, 2024
|
|
8RUY
 
 | |
8Y52
 
 | | Cryo-EM structure of the BA1-bound BRS3-Gq complex | | Descriptor: | BA1, Bombesin receptor subtype-3, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Li, C, Xu, Y, Yin, W, Xu, H.E. | | Deposit date: | 2024-01-31 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural insights into ligand recognition, selectivity, and activation of bombesin receptor subtype-3.
Cell Rep, 43, 2024
|
|
8Y53
 
 | | Cryo-EM structure of the MK-5046-bound BRS3-Gq complex | | Descriptor: | (2~{S})-1,1,1-tris(fluoranyl)-2-(4-pyrazol-1-ylphenyl)-3-[5-[[1-(trifluoromethyl)cyclopropyl]methyl]-1~{H}-imidazol-2-yl]propan-2-ol, Bombesin receptor subtype-3, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Li, C, Xu, Y, Yin, W, Xu, H.E. | | Deposit date: | 2024-01-31 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Structural insights into ligand recognition, selectivity, and activation of bombesin receptor subtype-3.
Cell Rep, 43, 2024
|
|
8Y5H
 
 | |
8Y5F
 
 | | Cryo-EM structure of E.coli spermidine transporter PotABC | | Descriptor: | Spermidine/putrescine import ATP-binding protein PotA, Spermidine/putrescine transport system permease protein PotB, Spermidine/putrescine transport system permease protein PotC | | Authors: | Qiao, Z, Gao, Y.G. | | Deposit date: | 2024-01-31 | | Release date: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Structural insights into polyamine spermidine uptake by the ABC transporter PotD-PotABC.
Sci Adv, 10, 2024
|
|
8Y5G
 
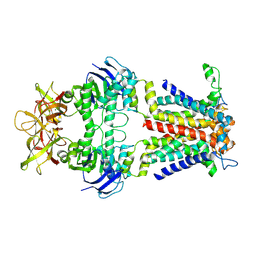 | |
8Y5I
 
 | |
8RV2
 
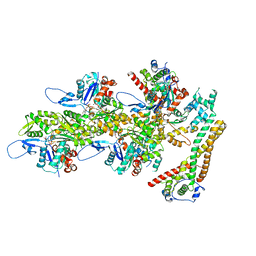 | | Structure of the formin INF2 bound to the barbed end of F-actin. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Actin, ... | | Authors: | Oosterheert, W, Boiero Sanders, M, Funk, J, Prumbaum, D, Raunser, S, Bieling, P. | | Deposit date: | 2024-01-31 | | Release date: | 2024-04-10 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Molecular mechanism of actin filament elongation by formins.
Science, 384, 2024
|
|
8RU8
 
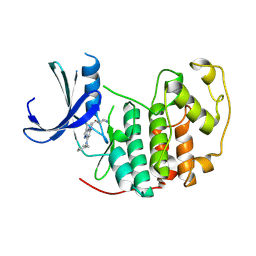 | | A crystal form of a human CDK2-CDK7 chimera | | Descriptor: | (3R,4R)-4-[[[7-[(phenylmethyl)amino]-3-propan-2-yl-pyrazolo[1,5-a]pyrimidin-5-yl]amino]methyl]piperidin-3-ol, Cyclin-dependent kinase 2 | | Authors: | Mukherjee, M, Cleasby, A. | | Deposit date: | 2024-01-30 | | Release date: | 2024-06-26 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Protein engineering enables a soakable crystal form of human CDK7 primed for high-throughput crystallography and structure-based drug design.
Structure, 32, 2024
|
|
8VUY
 
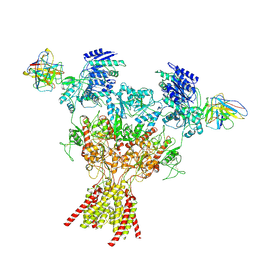 | | Rat GluN1-2B with Fab 003-102 | | Descriptor: | 003-102 Heavy, 003-102 Light, Glutamate receptor ionotropic, ... | | Authors: | Michalski, K, Furukawa, H. | | Deposit date: | 2024-01-30 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.81 Å) | | Cite: | Structural and functional mechanisms of anti-NMDAR autoimmune encephalitis.
Nat.Struct.Mol.Biol., 2024
|
|
