7PDZ
 
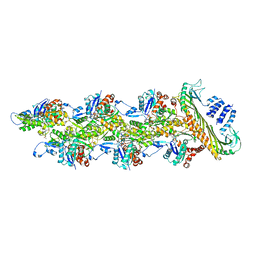 | | Structure of capping protein bound to the barbed end of a cytoplasmic actin filament | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, cytoplasmic 1, ... | | Authors: | Funk, J, Merino, F, Schacks, M, Rottner, K, Raunser, S, Bieling, P. | | Deposit date: | 2021-08-09 | | Release date: | 2021-09-01 | | Last modified: | 2021-10-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | A barbed end interference mechanism reveals how capping protein promotes nucleation in branched actin networks.
Nat Commun, 12, 2021
|
|
8RTY
 
 | | Structure of the F-actin barbed end bound by Cdc12 and profilin (ring complex) at a resolution of 6.3 Angstrom | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, cytoplasmic 1, ... | | Authors: | Oosterheert, W, Boiero Sanders, M, Funk, J, Prumbaum, D, Raunser, S, Bieling, P. | | Deposit date: | 2024-01-29 | | Release date: | 2024-04-10 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (6.25 Å) | | Cite: | Molecular mechanism of actin filament elongation by formins.
Science, 384, 2024
|
|
8RTT
 
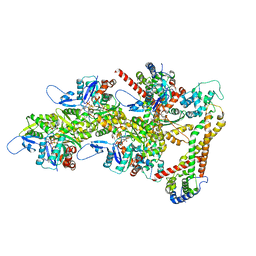 | | Structure of the formin Cdc12 bound to the barbed end of phalloidin-stabilized F-actin. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, cytoplasmic 1, ... | | Authors: | Oosterheert, W, Boiero Sanders, M, Funk, J, Prumbaum, D, Raunser, S, Bieling, P. | | Deposit date: | 2024-01-29 | | Release date: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.56 Å) | | Cite: | Mechanism of formin-mediated processive elongation of actin filaments revealed by cryo-EM
To Be Published
|
|
8RU0
 
 | | Structure of the undecorated barbed end of F-actin. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Actin, ... | | Authors: | Oosterheert, W, Boiero Sanders, M, Funk, J, Prumbaum, D, Raunser, S, Bieling, P. | | Deposit date: | 2024-01-29 | | Release date: | 2024-04-10 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Molecular mechanism of actin filament elongation by formins.
Science, 384, 2024
|
|
8RU2
 
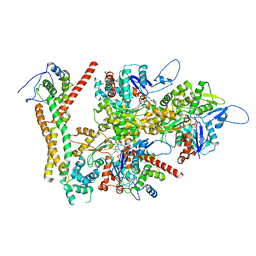 | | Structure of the F-actin barbed end bound by formin mDia1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, cytoplasmic 1, ... | | Authors: | Oosterheert, W, Boiero Sanders, M, Funk, J, Prumbaum, D, Raunser, S, Bieling, P. | | Deposit date: | 2024-01-29 | | Release date: | 2024-04-10 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Molecular mechanism of actin filament elongation by formins.
Science, 384, 2024
|
|
8RV2
 
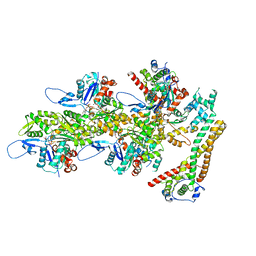 | | Structure of the formin INF2 bound to the barbed end of F-actin. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Actin, ... | | Authors: | Oosterheert, W, Boiero Sanders, M, Funk, J, Prumbaum, D, Raunser, S, Bieling, P. | | Deposit date: | 2024-01-31 | | Release date: | 2024-04-10 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Molecular mechanism of actin filament elongation by formins.
Science, 384, 2024
|
|
6FHL
 
 | | Cryo-EM structure of F-actin in complex with ADP-Pi | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Merino, F, Pospich, S, Funk, J, Wagner, T, Kuellmer, F, Arndt, H.-D, Bieling, P, Raunser, S. | | Deposit date: | 2018-01-15 | | Release date: | 2018-06-13 | | Last modified: | 2018-08-29 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural transitions of F-actin upon ATP hydrolysis at near-atomic resolution revealed by cryo-EM.
Nat. Struct. Mol. Biol., 25, 2018
|
|
7ZBQ
 
 | | Structure of the ADP-ribosyltransferase TccC3HVR from Photorhabdus Luminescens | | Descriptor: | TccC3 | | Authors: | Lindemann, F, Belyy, A, Friedrich, D, Schmieder, P, Bardiaux, B, Roderer, D, Funk, J, Protze, J, Bieling, P, Oschkinat, H, Raunser, S. | | Deposit date: | 2022-03-24 | | Release date: | 2022-06-29 | | Last modified: | 2022-08-03 | | Method: | SOLUTION NMR | | Cite: | Mechanism of threonine ADP-ribosylation of F-actin by a Tc toxin.
Nat Commun, 13, 2022
|
|
5OOE
 
 | | Cryo-EM structure of F-actin in complex with AppNHp (AMPPNP) | | Descriptor: | Actin, alpha skeletal muscle, MAGNESIUM ION, ... | | Authors: | Merino, F, Pospich, S, Funk, J, Kuellmer, F, Arndt, H.-D, Bieling, P, Raunser, S. | | Deposit date: | 2017-08-07 | | Release date: | 2018-06-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural transitions of F-actin upon ATP hydrolysis at near-atomic resolution revealed by cryo-EM.
Nat. Struct. Mol. Biol., 25, 2018
|
|
5OOD
 
 | | Cryo-EM structure of jasplakinolide-stabilized F-actin in complex with ADP-Pi | | Descriptor: | (4~{R},7~{R},10~{S},13~{S},15~{E},19~{S})-10-(4-azanylbutyl)-4-(4-hydroxyphenyl)-7-(1~{H}-indol-3-ylmethyl)-8,13,15,19-tetramethyl-1-oxa-5,8,11-triazacyclononadec-15-ene-2,6,9,12-tetrone, ADENOSINE-5'-DIPHOSPHATE, Actin, ... | | Authors: | Merino, F, Pospich, S, Funk, J, Kuellmer, F, Arndt, H.-D, Bieling, P, Raunser, S. | | Deposit date: | 2017-08-07 | | Release date: | 2018-06-13 | | Last modified: | 2019-12-04 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural transitions of F-actin upon ATP hydrolysis at near-atomic resolution revealed by cryo-EM.
Nat. Struct. Mol. Biol., 25, 2018
|
|
5ONV
 
 | | Cryo-EM structure of F-actin in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Merino, F, Pospich, S, Funk, J, Kuellmer, F, Arndt, H.-D, Bieling, P, Raunser, S. | | Deposit date: | 2017-08-04 | | Release date: | 2018-06-13 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural transitions of F-actin upon ATP hydrolysis at near-atomic resolution revealed by cryo-EM.
Nat. Struct. Mol. Biol., 25, 2018
|
|
5OOC
 
 | | Cryo-EM structure of jasplakinolide-stabilized F-actin in complex with ADP | | Descriptor: | (4~{R},7~{R},10~{S},13~{S},15~{E},19~{S})-10-(4-azanylbutyl)-4-(4-hydroxyphenyl)-7-(1~{H}-indol-3-ylmethyl)-8,13,15,19-tetramethyl-1-oxa-5,8,11-triazacyclononadec-15-ene-2,6,9,12-tetrone, ADENOSINE-5'-DIPHOSPHATE, Actin, ... | | Authors: | Merino, F, Pospich, S, Funk, J, Kuellmer, F, Arndt, H.-D, Bieling, P, Raunser, S. | | Deposit date: | 2017-08-07 | | Release date: | 2018-06-13 | | Last modified: | 2019-10-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural transitions of F-actin upon ATP hydrolysis at near-atomic resolution revealed by cryo-EM.
Nat. Struct. Mol. Biol., 25, 2018
|
|
5OOF
 
 | | Cryo-EM structure of F-actin in complex with ADP-BeFx | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Merino, F, Pospich, S, Funk, J, Kuellmer, F, Arndt, H.-D, Bieling, P, Raunser, S. | | Deposit date: | 2017-08-07 | | Release date: | 2018-06-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural transitions of F-actin upon ATP hydrolysis at near-atomic resolution revealed by cryo-EM.
Nat. Struct. Mol. Biol., 25, 2018
|
|
7ARE
 
 | | DNA origami pointer object v2 | | Descriptor: | SCAFFOLD STRAND, STAPLE STRAND | | Authors: | Thomas, M, Feigl, E, Kohler, F, Kube, M, Nagel-Yuksel, B, Willner, E.M, Funke, J.J, Gerling, T, Stommer, P, Honemann, M.N, Martin, T.G, Scheres, S.H.W, Dietz, H. | | Deposit date: | 2020-10-24 | | Release date: | 2020-11-18 | | Last modified: | 2020-12-30 | | Method: | ELECTRON MICROSCOPY (7.4 Å) | | Cite: | Revealing the structures of megadalton-scale DNA complexes with nucleotide resolution.
Nat Commun, 11, 2020
|
|
7AS5
 
 | | 126 helix bundle DNA nanostructure | | Descriptor: | SCAFFOLD STRAND, STAPLE STRAND | | Authors: | Kube, M, Kohler, F, Feigl, E, Nagel-Yuksel, B, Willner, E.M, Funke, J.J, Gerling, T, Stommer, P, Honemann, M.N, Martin, T.G, Scheres, S.H.W, Dietz, H. | | Deposit date: | 2020-10-27 | | Release date: | 2020-11-18 | | Last modified: | 2020-12-30 | | Method: | ELECTRON MICROSCOPY (9.8 Å) | | Cite: | Revealing the structures of megadalton-scale DNA complexes with nucleotide resolution.
Nat Commun, 11, 2020
|
|
7ART
 
 | | 48 helix bundle DNA origami brick | | Descriptor: | SCAFFOLD STRAND, STAPLE STRAND | | Authors: | Feigl, E, Kube, M, Kohler, F, Nagel-Yuksel, B, Willner, E.M, Funke, J.J, Gerling, T, Stommer, P, Honemann, M.N, Martin, T.G, Scheres, S.H.W, Dietz, H. | | Deposit date: | 2020-10-26 | | Release date: | 2020-11-11 | | Last modified: | 2020-12-30 | | Method: | ELECTRON MICROSCOPY (10 Å) | | Cite: | Revealing the structures of megadalton-scale DNA complexes with nucleotide resolution.
Nat Commun, 11, 2020
|
|
7ARV
 
 | | TwistTower_native-twist | | Descriptor: | SCAFFOLD STRAND, STAPLE STRAND | | Authors: | Kube, M, Kohler, F, Feigl, E, Nagel-Yuksel, B, Willner, E.M, Funke, J.J, Gerling, T, Stommer, P, Honemann, M.N, Martin, T.G, Scheres, S.H.W, Dietz, H. | | Deposit date: | 2020-10-26 | | Release date: | 2020-11-18 | | Last modified: | 2020-12-30 | | Method: | ELECTRON MICROSCOPY (7.4 Å) | | Cite: | Revealing the structures of megadalton-scale DNA complexes with nucleotide resolution.
Nat Commun, 11, 2020
|
|
7ARY
 
 | | Twist-Tower_twist-corrected-variant | | Descriptor: | SCAFFOLD STRAND, STAPLE STRAND | | Authors: | Kube, M, Kohler, F, Feigl, E, Nagel-Yuksel, B, Willner, E.M, Funke, J.J, Gerling, T, Stommer, P, Honemann, M.N, Martin, T.G, Scheres, S.H.W, Dietz, H. | | Deposit date: | 2020-10-26 | | Release date: | 2020-11-18 | | Last modified: | 2020-12-30 | | Method: | ELECTRON MICROSCOPY (8.5 Å) | | Cite: | Revealing the structures of megadalton-scale DNA complexes with nucleotide resolution.
Nat Commun, 11, 2020
|
|
