8G4V
 
 | | Horse liver alcohol dehydrogense His-51-Gln form complexed with NAD+ and 2,3,4,5,6-pentafluorobenzyl alcohol | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2,3,4,5,6-PENTAFLUOROBENZYL ALCOHOL, Alcohol dehydrogenase E chain, ... | | Authors: | Plapp, B.V, Subramanian, R. | | Deposit date: | 2023-02-10 | | Release date: | 2023-02-22 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Histidine-51 facilitates deprotonation of the zinc-bound ligand during catalysis by horse liver alcohol dehydrogenase
To Be Published
|
|
8G3E
 
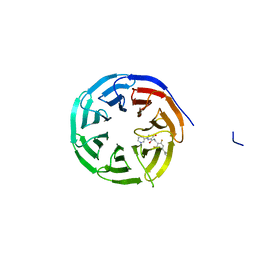 | | Crystal structure of human WDR5 in complex with (1M)-N-[(3,5-difluoro[1,1'-biphenyl]-4-yl)methyl]-6-methyl-4-oxo-1-(pyridin-3-yl)-1,4-dihydropyridazine-3-carboxamide (compound 2, WDR5-MYC inhibitor) | | Descriptor: | (1M)-N-[(3,5-difluoro[1,1'-biphenyl]-4-yl)methyl]-6-methyl-4-oxo-1-(pyridin-3-yl)-1,4-dihydropyridazine-3-carboxamide, WD repeat-containing protein 5 | | Authors: | Zhao, M. | | Deposit date: | 2023-02-07 | | Release date: | 2023-06-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Discovery and Structure-Based Design of Inhibitors of the WD Repeat-Containing Protein 5 (WDR5)-MYC Interaction.
J.Med.Chem., 66, 2023
|
|
8G3C
 
 | |
8G2L
 
 | |
8G2K
 
 | |
8G1U
 
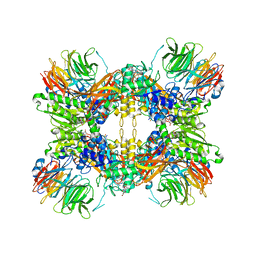 | | Structure of the methylosome-Lsm10/11 complex | | Descriptor: | ADENOSINE, Methylosome protein 50, Methylosome subunit pICln, ... | | Authors: | Lin, M, Paige, A, Tong, L. | | Deposit date: | 2023-02-03 | | Release date: | 2023-08-23 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | In vitro methylation of the U7 snRNP subunits Lsm11 and SmE by the PRMT5/MEP50/pICln methylosome.
Rna, 29, 2023
|
|
8G1F
 
 | | Structure of ACLY-D1026A-products | | Descriptor: | (3S)-citryl-Coenzyme A, ACETYL COENZYME *A, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Wei, X, Marmorstein, R. | | Deposit date: | 2023-02-02 | | Release date: | 2023-05-10 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Allosteric role of the citrate synthase homology domain of ATP citrate lyase.
Nat Commun, 14, 2023
|
|
8G1E
 
 | | Structure of ACLY-D1026A-products-asym | | Descriptor: | (3S)-citryl-Coenzyme A, ACETYL COENZYME *A, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Wei, X, Marmorstein, R. | | Deposit date: | 2023-02-02 | | Release date: | 2023-05-10 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Allosteric role of the citrate synthase homology domain of ATP citrate lyase.
Nat Commun, 14, 2023
|
|
8G0R
 
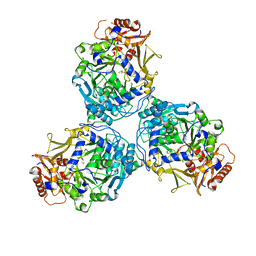 | |
8FYH
 
 | | G4 RNA-mediated PRC2 dimer | | Descriptor: | G4 RNA, Histone-binding protein RBBP4, Histone-lysine N-methyltransferase EZH2, ... | | Authors: | Song, J, Kasinath, V. | | Deposit date: | 2023-01-26 | | Release date: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for inactivation of PRC2 by G-quadruplex RNA.
Science, 381, 2023
|
|
8FXR
 
 | | Structure of neck with portal vertex of capsid of Agrobacterium phage Milano | | Descriptor: | Collar sheath protein, gp13, Linking protein 1, ... | | Authors: | Sonani, R.R, Wang, F, Esteves, N.C, Kelly, R.J, Sebastian, A, Kreutzberger, M.A.B, Leiman, P.G, Scharf, B.E, Egelman, E.H. | | Deposit date: | 2023-01-25 | | Release date: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Neck and capsid architecture of the robust Agrobacterium phage Milano.
Commun Biol, 6, 2023
|
|
8FXJ
 
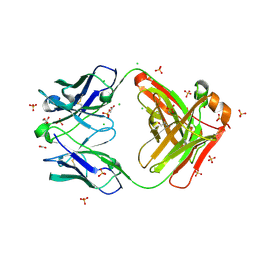 | | Crystal structure of Fab460 | | Descriptor: | ACETATE ION, CHLORIDE ION, Fab460, ... | | Authors: | Tan, K, Kim, M, Reinherz, E.L. | | Deposit date: | 2023-01-24 | | Release date: | 2023-10-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Inadequate structural constraint on Fab approach rather than paratope elicitation limits HIV-1 MPER vaccine utility.
Nat Commun, 14, 2023
|
|
8FWN
 
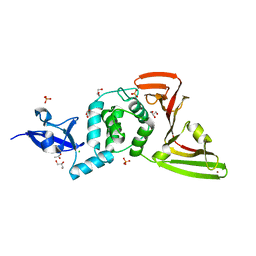 | | Crystal structure of SARS-CoV-2 papain-like protease C111S mutant | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Bezerra, E.H.S, Soprano, A.S, Tonoli, C.C.C, Prado, P.F.V, da Silva, J.C, Franchini, K.G, Trivella, D.B.B, Benedetti, C.E. | | Deposit date: | 2023-01-23 | | Release date: | 2023-05-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of SARS-CoV-2 papain-like protease C111S mutant
To Be Published
|
|
8FWG
 
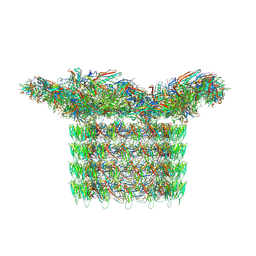 | | Structure of neck and portal vertex of Agrobacterium phage Milano, C5 symmetry | | Descriptor: | Collar sheath protein, gp13, Linking protein 1, ... | | Authors: | Sonani, R.R, Wang, F, Esteves, N.C, Kelly, R.J, Sebastian, A, Kreutzberger, M.A.B, Leiman, P.G, Scharf, B.E, Egelman, E.H. | | Deposit date: | 2023-01-22 | | Release date: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Neck and capsid architecture of the robust Agrobacterium phage Milano.
Commun Biol, 6, 2023
|
|
8FWF
 
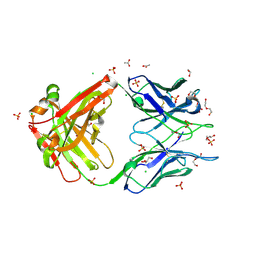 | | Crystal structure of Apo form Fab235 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Tan, K, Kim, M, Reinherz, E.L. | | Deposit date: | 2023-01-21 | | Release date: | 2023-10-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Inadequate structural constraint on Fab approach rather than paratope elicitation limits HIV-1 MPER vaccine utility.
Nat Commun, 14, 2023
|
|
8FUI
 
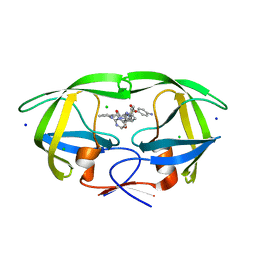 | | HIV-1 wild type protease with GRL-02519A, with N-(2,5-dimethylphenyl)-4-(pyridin-3-yl)pyrimidin-2-amine as P2-P3 group | | Descriptor: | ACETATE ION, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Wang, Y.-F, Wong-Sam, A.E, Ghosh, A.K, Weber, I.T. | | Deposit date: | 2023-01-17 | | Release date: | 2023-05-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Exploration of imatinib and nilotinib-derived templates as the P2-Ligand for HIV-1 protease inhibitors: Design, synthesis, protein X-ray structural studies, and biological evaluation.
Eur.J.Med.Chem., 255, 2023
|
|
8FR7
 
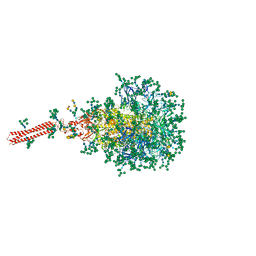 | | A hinge glycan regulates spike bending and impacts coronavirus infectivity | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-2)-[alpha-D-mannopyranose-(1-3)]alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pintilie, G, Wilson, E, Chmielewski, D, Schmid, M.F, Jin, J, Chen, M, Singharoy, A, Chiu, W. | | Deposit date: | 2023-01-06 | | Release date: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | A hinge glycan regulates spike bending and impacts coronavirus infectivity
To Be Published
|
|
8FOV
 
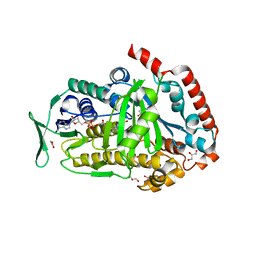 | | AbeH (Tryptophan-5-halogenase) bound to FAD and Cl | | Descriptor: | ACETATE ION, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Ashaduzzaman, M, Bellizzi, J.J. | | Deposit date: | 2023-01-03 | | Release date: | 2023-08-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystallographic and thermodynamic evidence of negative cooperativity of flavin and tryptophan binding in the flavin-dependent halogenases AbeH and BorH.
Biorxiv, 2023
|
|
8FJN
 
 | |
8FJM
 
 | |
8FGV
 
 | | Structure of rat neuronal nitric oxide synthase H692F mutant heme domain in complex with 6-(5-(2-(dimethylamino)ethyl)-2,3-difluorophenethyl)-4-methoxypyridin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6-(2-{5-[2-(dimethylamino)ethyl]-2,3-difluorophenyl}ethyl)-4-methoxypyridin-2-amine, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2022-12-12 | | Release date: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.848 Å) | | Cite: | Potent, Selective, and Membrane Permeable 2-Amino-4-Substituted Pyridine-Based Neuronal Nitric Oxide Synthase Inhibitors.
J.Med.Chem., 66, 2023
|
|
8FGE
 
 | | Structure of rat neuronal nitric oxide synthase R349A mutant heme domain in complex with 4-(difluoromethyl)-6-(5-(2-(dimethylamino)ethyl)-2,3-difluorophenethyl)pyridin-2-amine dihydrochloride | | Descriptor: | 4-(difluoromethyl)-6-(2-{5-[2-(dimethylamino)ethyl]-2,3-difluorophenyl}ethyl)pyridin-2-amine, 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2022-12-12 | | Release date: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Potent, Selective, and Membrane Permeable 2-Amino-4-Substituted Pyridine-Based Neuronal Nitric Oxide Synthase Inhibitors.
J.Med.Chem., 66, 2023
|
|
8FGD
 
 | | Structure of rat neuronal nitric oxide synthase R349A mutant heme domain in complex with 6-(5-(2-(diethylamino)ethyl)-2,3-difluorophenethyl)-4-methylpyridin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6-(2-{5-[2-(diethylamino)ethyl]-2,3-difluorophenyl}ethyl)-4-methylpyridin-2-amine, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2022-12-12 | | Release date: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.776 Å) | | Cite: | Potent, Selective, and Membrane Permeable 2-Amino-4-Substituted Pyridine-Based Neuronal Nitric Oxide Synthase Inhibitors.
J.Med.Chem., 66, 2023
|
|
8FGC
 
 | | Structure of rat neuronal nitric oxide synthase heme domain in complex with 6-(5-(2-aminoethyl)-2,3-difluorophenethyl)-4-methylpyridin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6-{2-[5-(2-aminoethyl)-2,3-difluorophenyl]ethyl}-4-methylpyridin-2-amine, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2022-12-12 | | Release date: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.779 Å) | | Cite: | Potent, Selective, and Membrane Permeable 2-Amino-4-Substituted Pyridine-Based Neuronal Nitric Oxide Synthase Inhibitors.
J.Med.Chem., 66, 2023
|
|
8FGB
 
 | | Structure of rat neuronal nitric oxide synthase heme domain in complex with 4-(5-(2-(dimethylamino)ethyl)-2,3-difluorophenethyl)-6-methylpyrimidin-2-amine | | Descriptor: | 4-(2-{5-[2-(dimethylamino)ethyl]-2,3-difluorophenyl}ethyl)-6-methylpyrimidin-2-amine, 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2022-12-12 | | Release date: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Potent, Selective, and Membrane Permeable 2-Amino-4-Substituted Pyridine-Based Neuronal Nitric Oxide Synthase Inhibitors.
J.Med.Chem., 66, 2023
|
|
