8RTY
 
 | | Structure of the F-actin barbed end bound by Cdc12 and profilin (ring complex) at a resolution of 6.3 Angstrom | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, cytoplasmic 1, ... | | Authors: | Oosterheert, W, Boiero Sanders, M, Funk, J, Prumbaum, D, Raunser, S, Bieling, P. | | Deposit date: | 2024-01-29 | | Release date: | 2024-04-10 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (6.25 Å) | | Cite: | Molecular mechanism of actin filament elongation by formins.
Science, 384, 2024
|
|
1LDK
 
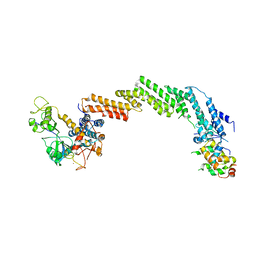 | | Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF Ubiquitin Ligase Complex | | Descriptor: | CULLIN HOMOLOG, CYCLIN A/CDK2-ASSOCIATED PROTEIN P19, SKP2-like protein type gamma, ... | | Authors: | Zheng, N, Schulman, B.A, Song, L, Miller, J.J, Jeffrey, P.D, Wang, P, Chu, C, Koepp, D.M, Elledge, S.J, Pagano, M, Conaway, R.C, Conaway, J.W, Harper, J.W, Pavletich, N.P. | | Deposit date: | 2002-04-08 | | Release date: | 2002-05-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex.
Nature, 416, 2002
|
|
8IHT
 
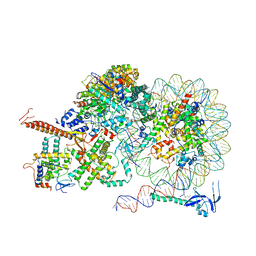 | | Rpd3S bound to the nucleosome | | Descriptor: | CALCIUM ION, Chromatin modification-related protein EAF3, DNA (164-MER), ... | | Authors: | Zhang, Y, Gang, C. | | Deposit date: | 2023-02-23 | | Release date: | 2024-03-20 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.72 Å) | | Cite: | Structural basis for nucleosome binding and catalysis by the yeast Rpd3S/HDAC holoenzyme.
Cell Res., 33, 2023
|
|
8JHO
 
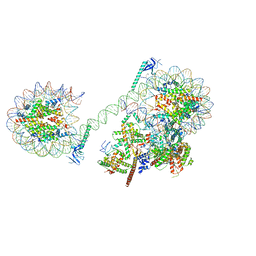 | |
6G0L
 
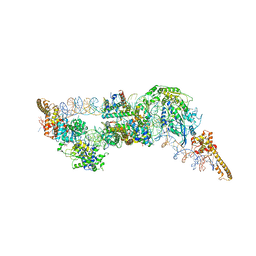 | | Structure of two molecules of the chromatin remodelling enzyme Chd1 bound to a nucleosome | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Chromo domain-containing protein 1, ... | | Authors: | Sundaramoorthy, R, Owen-hughes, T, Norman, D.G, Hughes, A. | | Deposit date: | 2018-03-19 | | Release date: | 2018-08-22 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (10 Å) | | Cite: | Structure of the chromatin remodelling enzyme Chd1 bound to a ubiquitinylated nucleosome.
Elife, 7, 2018
|
|
8R5H
 
 | | Ubiquitin ligation to neosubstrate by a cullin-RING E3 ligase & Cdc34: NEDD8-CUL2-RBX1-ELOB/C-VHL-MZ1 with trapped UBE2R2~donor UB-BRD4 BD2 | | Descriptor: | (2~{S},4~{R})-1-[(2~{S})-2-[2-[2-[2-[2-[2-[(9~{S})-7-(4-chlorophenyl)-4,5,13-trimethyl-3-thia-1,8,11,12-tetrazatricyclo[8.3.0.0^{2,6}]trideca-2(6),4,7,10,12-pentaen-9-yl]ethanoylamino]ethoxy]ethoxy]ethoxy]ethanoylamino]-3,3-dimethyl-butanoyl]-~{N}-[[4-(4-methyl-2,3-dihydro-1,3-thiazol-5-yl)phenyl]methyl]-4-oxidanyl-pyrrolidine-2-carboxamide, 5-azanylpentan-2-one, Bromodomain-containing protein 4, ... | | Authors: | Liwocha, J, Prabu, J.R, Kleiger, G, Schulman, B.A. | | Deposit date: | 2023-11-16 | | Release date: | 2024-02-21 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Cullin-RING ligases employ geometrically optimized catalytic partners for substrate targeting.
Mol.Cell, 84, 2024
|
|
8W9F
 
 | |
8W9E
 
 | |
8W9D
 
 | |
6R7F
 
 | | Structural basis of Cullin-2 RING E3 ligase regulation by the COP9 signalosome | | Descriptor: | COP9 signalosome complex subunit 1, COP9 signalosome complex subunit 2, COP9 signalosome complex subunit 3, ... | | Authors: | Faull, S.V, Lau, A.M.C, Martens, C, Ahdash, Z, Yebenes, H, Schmidt, C, Beuron, F, Cronin, N.B, Morris, E.P, Politis, A. | | Deposit date: | 2019-03-28 | | Release date: | 2019-08-28 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (8.2 Å) | | Cite: | Structural basis of Cullin 2 RING E3 ligase regulation by the COP9 signalosome.
Nat Commun, 10, 2019
|
|
6R7H
 
 | | Structural basis of Cullin-2 RING E3 ligase regulation by the COP9 signalosome | | Descriptor: | COP9 signalosome complex subunit 1, COP9 signalosome complex subunit 2, COP9 signalosome complex subunit 3, ... | | Authors: | Faull, S.V, Lau, A.M.C, Beuron, F, Cronin, N.B, Morris, E.P, Politis, A. | | Deposit date: | 2019-03-28 | | Release date: | 2019-08-28 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (8.8 Å) | | Cite: | Structural basis of Cullin 2 RING E3 ligase regulation by the COP9 signalosome.
Nat Commun, 10, 2019
|
|
8QZM
 
 | | Structure of DNMT3A1 UDR region bound to H2AK119ub nucleosome | | Descriptor: | DNA (145-MER), DNA (cytosine-5)-methyltransferase 3A, Histone H2A type 1, ... | | Authors: | Burdett, H, Wapenaar, H, Wilson, M.D. | | Deposit date: | 2023-10-27 | | Release date: | 2024-10-23 | | Last modified: | 2024-12-18 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The N-terminal region of DNMT3A engages the nucleosome surface to aid chromatin recruitment.
Embo Rep., 25, 2024
|
|
8X19
 
 | | Structure of nucleosome-bound SRCAP-C in the ADP-BeFx-bound state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Actin, ... | | Authors: | Yu, J, Wang, Q, Yu, Z, Li, W, Wang, L, Xu, Y. | | Deposit date: | 2023-11-06 | | Release date: | 2024-03-06 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into histone exchange by human SRCAP complex.
Cell Discov, 10, 2024
|
|
8X15
 
 | | Structure of nucleosome-bound SRCAP-C in the apo state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Actin, ... | | Authors: | Yu, J, Wang, Q, Yu, Z, Li, W, Wang, L, Xu, Y. | | Deposit date: | 2023-11-06 | | Release date: | 2024-03-06 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into histone exchange by human SRCAP complex.
Cell Discov, 10, 2024
|
|
8X1C
 
 | | Structure of nucleosome-bound SRCAP-C in the ADP-bound state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Actin, ... | | Authors: | Yu, J, Wang, Q, Yu, Z, Li, W, Wang, L, Xu, Y. | | Deposit date: | 2023-11-06 | | Release date: | 2024-03-06 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into histone exchange by human SRCAP complex.
Cell Discov, 10, 2024
|
|
7K7G
 
 | | nucleosome and Gal4 complex | | Descriptor: | Centromere DNA-binding protein complex CBF3 subunit B, DNA (147-MER), Histone H2A.1, ... | | Authors: | Ruifang, G, Yawen, B. | | Deposit date: | 2020-09-22 | | Release date: | 2021-03-31 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural and dynamic mechanisms of CBF3-guided centromeric nucleosome formation.
Nat Commun, 12, 2021
|
|
7X3T
 
 | | Cryo-EM structure of ISW1a-dinucleosome | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DNA (343-MER), ... | | Authors: | Lifei, L, Kangjing, C, Chen, Z. | | Deposit date: | 2022-03-01 | | Release date: | 2023-09-20 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (5.4 Å) | | Cite: | Structure of the ISW1a complex bound to the dinucleosome.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8IJ1
 
 | | Protomer 1 and 2 of the asymmetry trimer of the Cul2-Rbx1-EloBC-FEM1B ubiquitin ligase complex | | Descriptor: | Cullin-2, E3 ubiquitin-protein ligase RBX1, Elongin-B, ... | | Authors: | Dai, Z, Liang, L, Yin, Y.X. | | Deposit date: | 2023-02-24 | | Release date: | 2024-02-28 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural insights into the ubiquitylation strategy of the oligomeric CRL2 FEM1B E3 ubiquitin ligase.
Embo J., 43, 2024
|
|
5KC2
 
 | | Negative stain structure of Vps15/Vps34 complex | | Descriptor: | Phosphatidylinositol 3-kinase VPS34, Serine/threonine-protein kinase VPS15 | | Authors: | Kirsten, M.L, Zhang, L, Ohashi, Y, Perisic, O, Williams, R.L, Sachse, C. | | Deposit date: | 2016-06-04 | | Release date: | 2016-10-05 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (28 Å) | | Cite: | Characterization of Atg38 and NRBF2, a fifth subunit of the autophagic Vps34/PIK3C3 complex.
Autophagy, 12, 2016
|
|
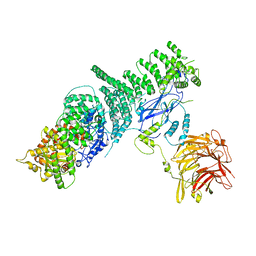
3DZU
 
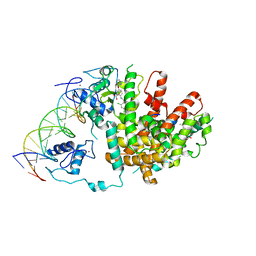 | | Intact PPAR gamma - RXR alpha Nuclear Receptor Complex on DNA bound with BVT.13, 9-cis Retinoic Acid and NCOA2 Peptide | | Descriptor: | (9cis)-retinoic acid, 2-[(2,4-DICHLOROBENZOYL)AMINO]-5-(PYRIMIDIN-2-YLOXY)BENZOIC ACID, DNA (5'-D(*DCP*DAP*DAP*DAP*DCP*DTP*DAP*DGP*DGP*DTP*DCP*DAP*DAP*DAP*DGP*DGP*DTP*DCP*DAP*DG)-3'), ... | | Authors: | Chandra, V, Huang, P, Hamuro, Y, Raghuram, S, Wang, Y, Burris, T.P, Rastinejad, F. | | Deposit date: | 2008-07-30 | | Release date: | 2008-10-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of the intact PPAR-gamma-RXR- nuclear receptor complex on DNA.
Nature, 456, 2008
|
|
3E00
 
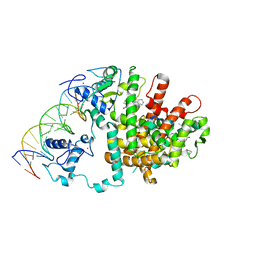 | | Intact PPAR gamma - RXR alpha Nuclear Receptor Complex on DNA bound with GW9662, 9-cis Retinoic Acid and NCOA2 Peptide | | Descriptor: | (9cis)-retinoic acid, 2-chloro-5-nitro-N-phenylbenzamide, DNA (5'-D(*DCP*DAP*DAP*DAP*DCP*DTP*DAP*DGP*DGP*DTP*DCP*DAP*DAP*DAP*DGP*DGP*DTP*DCP*DAP*DG)-3'), ... | | Authors: | Chandra, V, Huang, P, Hamuro, Y, Raghuram, S, Wang, Y, Burris, T.P, Rastinejad, F. | | Deposit date: | 2008-07-30 | | Release date: | 2008-10-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of the intact PPAR-gamma-RXR- nuclear receptor complex on DNA.
Nature, 456, 2008
|
|
3DZY
 
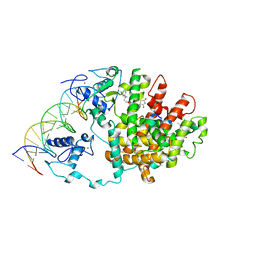 | | Intact PPAR gamma - RXR alpha Nuclear Receptor Complex on DNA bound with Rosiglitazone, 9-cis Retinoic Acid and NCOA2 Peptide | | Descriptor: | (9cis)-retinoic acid, 2,4-THIAZOLIDIINEDIONE, 5-[[4-[2-(METHYL-2-PYRIDINYLAMINO)ETHOXY]PHENYL]METHYL]-(9CL), ... | | Authors: | Chandra, V, Huang, P, Hamuro, Y, Raghuram, S, Wang, Y, Burris, T.P, Rastinejad, F. | | Deposit date: | 2008-07-30 | | Release date: | 2008-10-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of the intact PPAR-gamma-RXR- nuclear receptor complex on DNA.
Nature, 456, 2008
|
|
7AOA
 
 | | Structure of the extended MTA1/HDAC1/MBD2/RBBP4 NURD deacetylase complex | | Descriptor: | Histone deacetylase 1, Histone-binding protein RBBP4, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Millard, C.J, Fairall, L, Ragan, T.J, Savva, C.G, Schwabe, J.W.R. | | Deposit date: | 2020-10-14 | | Release date: | 2020-11-11 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (19.4 Å) | | Cite: | The topology of chromatin-binding domains in the NuRD deacetylase complex.
Nucleic Acids Res., 48, 2020
|
|
8KHP
 
 | | CULLIN3-KLHL22-RBX1 E3 ligase | | Descriptor: | Cullin-3, E3 ubiquitin-protein ligase RBX1, Kelch-like protein 22 | | Authors: | Su, M.-Y, Su, M.-Y. | | Deposit date: | 2023-08-22 | | Release date: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | Cryo-EM structure of the KLHL22 E3 ligase bound to an oligomeric metabolic enzyme.
Structure, 31, 2023
|
|
8CVT
 
 | | Human 19S-20S proteasome, state SD2 | | Descriptor: | 26S protease regulatory subunit 10B, 26S protease regulatory subunit 8, 26S proteasome complex subunit SEM1, ... | | Authors: | Zhao, J. | | Deposit date: | 2022-05-18 | | Release date: | 2022-11-02 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into the human PA28-20S proteasome enabled by efficient tagging and purification of endogenous proteins.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
