7VVM
 
 | | PTH-bound human PTH1R in complex with Gs (class3) | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Kobayashi, K, Kusakizako, T, Miyauchi, H, Tomita, A, Kobayashi, K, Shihoya, W, Yamashita, K, Nishizawa, T, Kato, H.E, Nureki, O. | | Deposit date: | 2021-11-06 | | Release date: | 2022-08-03 | | Last modified: | 2023-02-15 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Endogenous ligand recognition and structural transition of a human PTH receptor.
Mol.Cell, 82, 2022
|
|
7VVO
 
 | | PTH-bound human PTH1R in complex with Gs (class5) | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Kobayashi, K, Kusakizako, T, Miyauchi, H, Tomita, A, Kobayashi, K, Shihoya, W, Yamashita, K, Nishizawa, T, Kato, H.E, Nureki, O. | | Deposit date: | 2021-11-06 | | Release date: | 2022-08-03 | | Last modified: | 2023-02-15 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Endogenous ligand recognition and structural transition of a human PTH receptor.
Mol.Cell, 82, 2022
|
|
6VAV
 
 | | Peanut lectin complexed with divalent N-beta-D-galactopyranosyl-L-succinamoyl derivative (diNGS) | | Descriptor: | CALCIUM ION, Galactose-binding lectin, MANGANESE (II) ION, ... | | Authors: | Otero, L.H, Primo, E.D, Cagnoni, A.J, Klinke, S, Goldbaum, F.A, Uhrig, M.L. | | Deposit date: | 2019-12-18 | | Release date: | 2020-10-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structures of peanut lectin in the presence of synthetic beta-N- and beta-S-galactosides disclose evidence for the recognition of different glycomimetic ligands.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6VC3
 
 | | Peanut lectin complexed with S-beta-D-thiogalactopyranosyl 6-deoxy-6-S-propynyl-beta-D-glucopyranoside (STG) | | Descriptor: | 6-S-(prop-2-yn-1-yl)-6-thio-beta-D-glucopyranosyl 1-thio-beta-D-galactopyranoside, CALCIUM ION, Galactose-binding lectin, ... | | Authors: | Otero, L.H, Primo, E.D, Cagnoni, A.J, Cano, M.E, Klinke, S, Goldbaum, F.A, Uhrig, M.L. | | Deposit date: | 2019-12-20 | | Release date: | 2020-10-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structures of peanut lectin in the presence of synthetic beta-N- and beta-S-galactosides disclose evidence for the recognition of different glycomimetic ligands.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
5J5X
 
 | | Complex of PKA with the bisubstrate protein kinase inhibitor ARC-1416 | | Descriptor: | 4-(piperazin-1-yl)-7H-pyrrolo[2,3-d]pyrimidine, 47P-AZ1-DAL-DAR-DAR-DAR-DAR, SULFATE ION, ... | | Authors: | Alam, K.A, Ivan, T, Uri, A, Engh, R.A. | | Deposit date: | 2016-04-04 | | Release date: | 2016-07-20 | | Last modified: | 2019-06-19 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Bifunctional Ligands for Inhibition of Tight-Binding Protein-Protein Interactions.
Bioconjug.Chem., 27, 2016
|
|
7C6R
 
 | | Crystal structure of beta-glycosides-binding protein (W177X) of ABC transporter in a closed state bound to cellopentaose | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, SULFATE ION, ... | | Authors: | Kanaujia, S.P, Chandravanshi, M, Samanta, R. | | Deposit date: | 2020-05-22 | | Release date: | 2020-09-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Conformational Trapping of a beta-Glucosides-Binding Protein Unveils the Selective Two-Step Ligand-Binding Mechanism of ABC Importers.
J.Mol.Biol., 432, 2020
|
|
7C68
 
 | | Crystal structure of beta-glycosides-binding protein of ABC transporter in a closed state bound to cellotetraose | | Descriptor: | 1,2-ETHANEDIOL, CARBON DIOXIDE, CHLORIDE ION, ... | | Authors: | Kanaujia, S.P, Chandravanshi, M, Samanta, R. | | Deposit date: | 2020-05-21 | | Release date: | 2020-09-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Conformational Trapping of a beta-Glucosides-Binding Protein Unveils the Selective Two-Step Ligand-Binding Mechanism of ABC Importers.
J.Mol.Biol., 432, 2020
|
|
7C69
 
 | | Crystal structure of beta-glycosides-binding protein of ABC transporter in a closed state bound to sophorose | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Sugar ABC transporter, ... | | Authors: | Kanaujia, S.P, Chandravanshi, M, Samanta, R. | | Deposit date: | 2020-05-21 | | Release date: | 2020-09-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Conformational Trapping of a beta-Glucosides-Binding Protein Unveils the Selective Two-Step Ligand-Binding Mechanism of ABC Importers.
J.Mol.Biol., 432, 2020
|
|
7C6V
 
 | | Crystal structure of beta-glycosides-binding protein (W177X) of ABC transporter in a closed state bound to laminaritriose (Form II) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kanaujia, S.P, Chandravanshi, M, Samanta, R. | | Deposit date: | 2020-05-22 | | Release date: | 2020-09-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Conformational Trapping of a beta-Glucosides-Binding Protein Unveils the Selective Two-Step Ligand-Binding Mechanism of ABC Importers.
J.Mol.Biol., 432, 2020
|
|
2XV3
 
 | | Pseudomonas aeruginosa Azurin with mutated metal-binding loop sequence (CAAAAHAAAAM), chemically reduced, pH5.3 | | Descriptor: | AZURIN, COPPER (I) ION | | Authors: | Li, C, Sato, K, Monari, S, Salard, I, Sola, M, Banfield, M.J, Dennison, C. | | Deposit date: | 2010-10-22 | | Release date: | 2010-12-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Metal-Binding Loop Length is a Determinant of the Pka of a Histidine Ligand at a Type 1 Copper Site
Inorg.Chem., 50, 2011
|
|
5UK5
 
 | | Complex of Notch1(EGF8-12) bound to Jagged1(N-EGF3) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Garcia, K.C, Luca, V.C. | | Deposit date: | 2017-01-19 | | Release date: | 2017-03-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.506 Å) | | Cite: | Notch-Jagged complex structure implicates a catch bond in tuning ligand sensitivity.
Science, 355, 2017
|
|
7ZXW
 
 | | Catalytic domain of UDP-Glucose Glycoprotein Glucosyltransferase from Chaetomium thermophilum in complex with the 5-[(morpholin-4-yl)methyl]quinolin-8-ol inhibitor | | Descriptor: | 5-(morpholin-4-ylmethyl)quinolin-8-ol, CALCIUM ION, DIMETHYL SULFOXIDE, ... | | Authors: | Le Cornu, J.D, Ibba, R, Roversi, P, Zitzmann, N. | | Deposit date: | 2022-05-23 | | Release date: | 2022-11-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.246 Å) | | Cite: | Crystal polymorphism in fragment-based lead discovery of ligands of the catalytic domain of UGGT, the glycoprotein folding quality control checkpoint.
Front Mol Biosci, 9, 2022
|
|
6SPV
 
 | | Crystal structure of PDZ1-2 from PSD-95 | | Descriptor: | Disks large homolog 4, GLUTATHIONE | | Authors: | Rodzli, N, Levy, C.W, Prince, S.M. | | Deposit date: | 2019-09-02 | | Release date: | 2019-10-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | The Dual PDZ Domain from Postsynaptic Density Protein 95 Forms a Scaffold with Peptide Ligand.
Biophys.J., 119, 2020
|
|
1KWT
 
 | | Rat mannose binding protein A (native, MPD) | | Descriptor: | CALCIUM ION, CHLORIDE ION, MANNOSE-BINDING PROTEIN A | | Authors: | Ng, K.K.S, Kolatkar, A.R, Park-Snyder, S, Feinberg, H, Clark, D.A, Drickamer, K, Weis, W.I. | | Deposit date: | 2002-01-30 | | Release date: | 2002-07-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Orientation of bound ligands in mannose-binding proteins. Implications for multivalent ligand recognition.
J.Biol.Chem., 277, 2002
|
|
1KWY
 
 | | Rat mannose protein A complexed with man-a13-man. | | Descriptor: | CALCIUM ION, CHLORIDE ION, MANNOSE-BINDING PROTEIN A, ... | | Authors: | Ng, K.K, Kolatkar, A.R, Park-Snyder, S, Feinberg, H, Clark, D.A, Drickamer, K, Weis, W.I. | | Deposit date: | 2002-01-30 | | Release date: | 2002-07-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Orientation of bound ligands in mannose-binding proteins. Implications for multivalent ligand recognition.
J.Biol.Chem., 277, 2002
|
|
4QZS
 
 | | Crystal structure of the first bromodomain of human 3-fluoro tyrosine-labeled brd4 in complex with jq1 | | Descriptor: | (6S)-6-(2-tert-butoxy-2-oxoethyl)-4-(4-chlorophenyl)-2,3,9-trimethyl-6,7-dihydrothieno[3,2-f][1,2,4]triazolo[4,3-a][1,4]diazepin-10-ium, 1,2-ETHANEDIOL, Bromodomain-containing protein 4, ... | | Authors: | Ember, S.W, Schonbrunn, E. | | Deposit date: | 2014-07-28 | | Release date: | 2014-10-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Fluorinated aromatic amino acids are sensitive (19)f NMR probes for bromodomain-ligand interactions.
Acs Chem.Biol., 9, 2014
|
|
5UT9
 
 | | Sperm whale myoglobin H64A with nitrite | | Descriptor: | GLYCEROL, Myoglobin, NITRITE ION, ... | | Authors: | Wang, B, Thomas, L.M, Richter-Addo, G.B. | | Deposit date: | 2017-02-14 | | Release date: | 2018-02-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Nitrosyl Myoglobins and Their Nitrite Precursors: Crystal Structural and Quantum Mechanics and Molecular Mechanics Theoretical Investigations of Preferred Fe -NO Ligand Orientations in Myoglobin Distal Pockets.
Biochemistry, 57, 2018
|
|
8I07
 
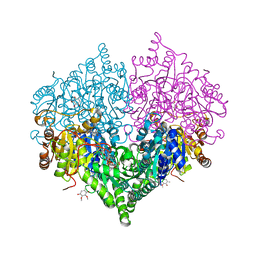 | |
8I08
 
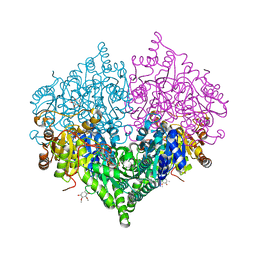 | | Crystal structure of Escherichia coli glyoxylate carboligase quadruple mutant | | Descriptor: | 2,3-DIMETHOXY-5-METHYL-1,4-BENZOQUINONE, FLAVIN-ADENINE DINUCLEOTIDE, Glyoxylate carboligase, ... | | Authors: | Kim, J.H, Kim, J.S. | | Deposit date: | 2023-01-10 | | Release date: | 2023-11-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Engineering of two thiamine diphosphate-dependent enzymes for the regioselective condensation of C1-formaldehyde into C4-erythrulose.
Int.J.Biol.Macromol., 253, 2023
|
|
8I01
 
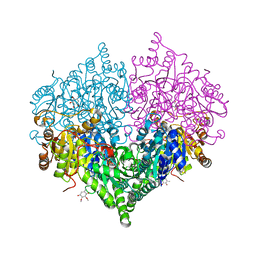 | | Crystal structure of Escherichia coli glyoxylate carboligase | | Descriptor: | 2,3-DIMETHOXY-5-METHYL-1,4-BENZOQUINONE, FLAVIN-ADENINE DINUCLEOTIDE, Glyoxylate carboligase, ... | | Authors: | Kim, J.H, Kim, J.S. | | Deposit date: | 2023-01-10 | | Release date: | 2023-11-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Engineering of two thiamine diphosphate-dependent enzymes for the regioselective condensation of C1-formaldehyde into C4-erythrulose.
Int.J.Biol.Macromol., 253, 2023
|
|
6CBQ
 
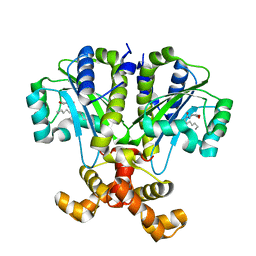 | | Crystal structure of QscR bound to agonist S3 | | Descriptor: | (2S)-2-hexyl-N-[(3S)-2-oxooxolan-3-yl]decanamide, LuxR family transcriptional regulator | | Authors: | Churchill, M.E.A, Wysoczynski-Horita, C.L. | | Deposit date: | 2018-02-05 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mechanism of agonism and antagonism of the Pseudomonas aeruginosa quorum sensing regulator QscR with non-native ligands.
Mol. Microbiol., 108, 2018
|
|
8I05
 
 | | Crystal structure of Escherichia coli glyoxylate carboligase double mutant | | Descriptor: | 2,3-DIMETHOXY-5-METHYL-1,4-BENZOQUINONE, FLAVIN-ADENINE DINUCLEOTIDE, Glyoxylate carboligase, ... | | Authors: | Kim, J.H, Kim, J.S. | | Deposit date: | 2023-01-10 | | Release date: | 2023-11-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Engineering of two thiamine diphosphate-dependent enzymes for the regioselective condensation of C1-formaldehyde into C4-erythrulose.
Int.J.Biol.Macromol., 253, 2023
|
|
6WJH
 
 | |
5V6P
 
 | | CryoEM structure of the ERAD-associated E3 ubiquitin-protein ligase HRD1 | | Descriptor: | ERAD-associated E3 ubiquitin-protein ligase HRD1 | | Authors: | Schoebel, S, Mi, W, Stein, A, Rapoport, T.A, Liao, M. | | Deposit date: | 2017-03-17 | | Release date: | 2017-08-16 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM structure of the protein-conducting ERAD channel Hrd1 in complex with Hrd3.
Nature, 548, 2017
|
|
7B6W
 
 | | Crystal structure of the human alpha1B adrenergic receptor in complex with inverse agonist (+)-cyclazosin | | Descriptor: | Alpha-1B adrenergic receptor,alpha1B adrenergic receptor,Alpha-1B adrenergic receptor,alpha1B adrenergic receptor,Alpha-1B adrenergic receptor,alpha1B adrenergic receptor,Alpha-1B adrenergic receptor,alpha1B adrenergic receptor, [(4~{a}~{R},8~{a}~{S})-4-(4-azanyl-6,7-dimethoxy-quinazolin-2-yl)-2,3,4~{a},5,6,7,8,8~{a}-octahydroquinoxalin-1-yl]-(furan-2-yl)methanone | | Authors: | Deluigi, M, Morstein, L, Hilge, M, Schuster, M, Merklinger, L, Klipp, A, Scott, D.J, Plueckthun, A. | | Deposit date: | 2020-12-08 | | Release date: | 2022-01-12 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.873 Å) | | Cite: | Crystal structure of the alpha 1B -adrenergic receptor reveals molecular determinants of selective ligand recognition.
Nat Commun, 13, 2022
|
|
