1N36
 
 | | Structure of the Thermus thermophilus 30S ribosomal subunit in the presence of crystallographically disordered codon and near-cognate transfer RNA anticodon stem-loop mismatched at the second codon position | | Descriptor: | 16S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Ogle, J.M, Murphy IV, F.V, Tarry, M.J, Ramakrishnan, V. | | Deposit date: | 2002-10-25 | | Release date: | 2002-11-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | Selection of tRNA by the Ribosome Requires a Transition from an Open to a Closed Form
Cell(Cambridge,Mass.), 111, 2002
|
|
5OHO
 
 | |
1R3F
 
 | | Crystal Structure of tRNA Pseudouridine Synthase TruB and Its RNA Complex: RNA-protein Recognition Through a Combination of Rigid Docking and Induced Fit | | Descriptor: | tRNA pseudouridine synthase B | | Authors: | Pan, H, Agarwalla, S, Moustakas, D.T, Finer-Moore, J, Stroud, R.M. | | Deposit date: | 2003-10-01 | | Release date: | 2003-11-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of tRNA Pseudouridine Synthase TruB and Its RNA Complex: RNA Recognition Through a Combination of Rigid Docking and Induced Fit
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
5OHQ
 
 | |
6SDY
 
 | |
4U3O
 
 | | Octameric RNA duplex soaked in manganese(II)chloride | | Descriptor: | MANGANESE (II) ION, RNA (5'-R(*UP*CP*GP*UP*AP*CP*GP*A)-3') | | Authors: | Schaffer, M.F, Spingler, B, Schnabl, J, Peng, G, Olieric, V, Sigel, R.K.O. | | Deposit date: | 2014-07-22 | | Release date: | 2015-07-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The X-ray Structures of Six Octameric RNA Duplexes in the Presence of Different Di- and Trivalent Cations.
Int J Mol Sci, 17, 2016
|
|
8FIY
 
 | | Cryo-EM structure of E. coli RNA polymerase Elongation complex in the Transcription-Translation Complex (RNAP in an anti-swiveled conformation) | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Florez Ariza, A, Wee, L, Tong, A, Canari, C, Grob, P, Nogales, E, Bustamante, C. | | Deposit date: | 2022-12-17 | | Release date: | 2023-03-29 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (7.3 Å) | | Cite: | A trailing ribosome speeds up RNA polymerase at the expense of transcript fidelity via force and allostery.
Cell, 186, 2023
|
|
4U3L
 
 | | octameric RNA duplex co-crystallized in calcium(II)chloride | | Descriptor: | CALCIUM ION, RNA (5'-R(*UP*CP*GP*UP*AP*CP*GP*A)-3') | | Authors: | Schaffer, M.F, Spingler, B, Schnabl, J, Peng, G, Olieric, V, Sigel, R.K.O. | | Deposit date: | 2014-07-22 | | Release date: | 2015-07-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.482 Å) | | Cite: | The X-ray Structures of Six Octameric RNA Duplexes in the Presence of Different Di- and Trivalent Cations.
Int J Mol Sci, 17, 2016
|
|
4U78
 
 | | Octameric RNA duplex soaked in copper(II)chloride | | Descriptor: | CALCIUM ION, COPPER (II) ION, RNA (5'-R(*UP*CP*GP*UP*AP*CP*GP*A)-3') | | Authors: | Schaffer, M.F, Spingler, B, Schnabl, J, Peng, G, Olieric, V, Sigel, R.K.O. | | Deposit date: | 2014-07-30 | | Release date: | 2015-08-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | The X-ray Structures of Six Octameric RNA Duplexes in the Presence of Different Di- and Trivalent Cations.
Int J Mol Sci, 17, 2016
|
|
7AHO
 
 | | RUVBL1-RUVBL2 heterohexameric ring after binding of RNA helicase DHX34 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, RuvB-like 1, RuvB-like 2 | | Authors: | Lopez-Perrote, A, Rodriguez, C.F, Llorca, O. | | Deposit date: | 2020-09-25 | | Release date: | 2020-11-25 | | Method: | ELECTRON MICROSCOPY (4.18 Å) | | Cite: | Regulation of RUVBL1-RUVBL2 AAA-ATPases by the nonsense-mediated mRNA decay factor DHX34, as evidenced by Cryo-EM.
Elife, 9, 2020
|
|
4U3R
 
 | | Octameric RNA duplex co-crystallized with cobalt(II)chloride | | Descriptor: | COBALT (II) ION, RNA (5'-R(*UP*CP*GP*UP*AP*CP*GP*A)-3') | | Authors: | Schaffer, M.F, Spingler, B, Schnabl, J, Peng, G, Olieric, V, Sigel, R.K.O. | | Deposit date: | 2014-07-22 | | Release date: | 2015-07-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | The X-ray Structures of Six Octameric RNA Duplexes in the Presence of Different Di- and Trivalent Cations.
Int J Mol Sci, 17, 2016
|
|
4PZ6
 
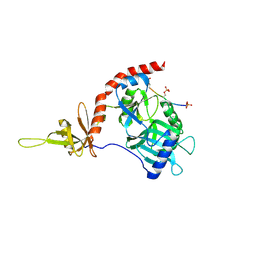 | |
1PN7
 
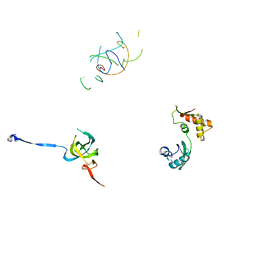 | | Coordinates of S12, L11 proteins and P-tRNA, from the 70S X-ray structure aligned to the 70S Cryo-EM map of E.coli ribosome | | Descriptor: | 30S ribosomal protein S12, 50S ribosomal protein L11, P-tRNA | | Authors: | Valle, M, Zavialov, A, Sengupta, J, Rawat, U, Ehrenberg, M, Frank, J. | | Deposit date: | 2003-06-12 | | Release date: | 2003-07-15 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (10.8 Å) | | Cite: | Locking and Unlocking of Ribosomal Motions
Cell(Cambridge,Mass.), 114, 2003
|
|
6PUN
 
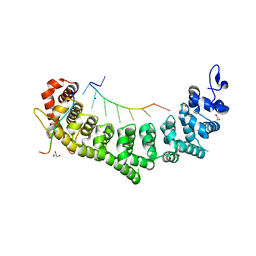 | | Crystal structure of a ternary complex of FBF-2 with LST-1 (site B) and compact FBE RNA | | Descriptor: | 1,2-ETHANEDIOL, Fem-3 mRNA-binding factor 2, GLYCEROL, ... | | Authors: | Qiu, C, Campbell, Z.T, Hall, T.M.T. | | Deposit date: | 2019-07-18 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | A crystal structure of a collaborative RNA regulatory complex reveals mechanisms to refine target specificity.
Elife, 8, 2019
|
|
6JP6
 
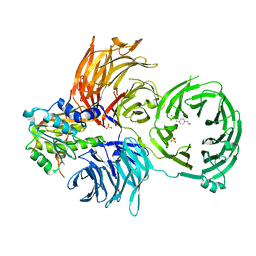 | | The X-ray structure of yeast tRNA methyltransferase complex of Trm7 and Trm734 essential for 2'-O-methylation at the first position of anticodon in specific tRNAs | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, SULFATE ION, tRNA (cytidine(34)/guanosine(34)-2'-O)-methyltransferase, ... | | Authors: | Hirata, A, Okada, K, Yoshii, K, Shiraisi, H, Saijo, S, Yonezawa, K, Sihimzu, N, Hori, H. | | Deposit date: | 2019-03-26 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.699 Å) | | Cite: | Structure of tRNA methyltransferase complex of Trm7 and Trm734 reveals a novel binding interface for tRNA recognition.
Nucleic Acids Res., 47, 2019
|
|
6NY3
 
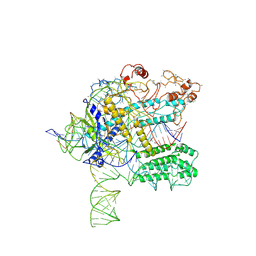 | | CasX ternary complex with 30bp target DNA | | Descriptor: | CasX, DNA Non-target strand, DNA target strand, ... | | Authors: | Liu, J.J, Orlova, N, Nogales, E, Doudna, J.A. | | Deposit date: | 2019-02-10 | | Release date: | 2019-02-27 | | Last modified: | 2019-12-25 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | CasX enzymes comprise a distinct family of RNA-guided genome editors.
Nature, 566, 2019
|
|
6O5F
 
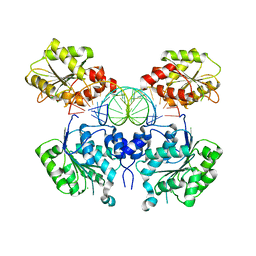 | | Crystal structure of DEAD-box RNA helicase DDX3X at pre-unwound state | | Descriptor: | ATP-dependent RNA helicase DDX3X, CHLORIDE ION, RNA (5'-R(P*CP*AP*AP*GP*GP*UP*CP*AP*UP*UP*CP*GP*CP*AP*AP*GP*AP*GP*UP*GP*GP*CP*C)-3') | | Authors: | Song, H, Ji, X. | | Deposit date: | 2019-03-02 | | Release date: | 2019-07-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.504 Å) | | Cite: | The mechanism of RNA duplex recognition and unwinding by DEAD-box helicase DDX3X.
Nat Commun, 10, 2019
|
|
8SQK
 
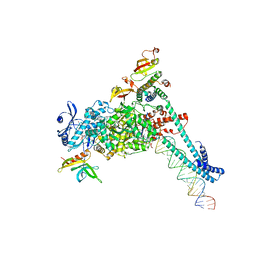 | | SARS-CoV-2 replication-transcription complex bound to RNA-nsp9 and GDP-betaS, as a pre-catalytic deRNAylation/mRNA capping intermediate | | Descriptor: | 5'-O-[(R)-hydroxy(thiophosphonooxy)phosphoryl]guanosine, MAGNESIUM ION, Non-structural protein 7, ... | | Authors: | Small, G.I, Darst, S.A, Campbell, E.A. | | Deposit date: | 2023-05-04 | | Release date: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structural and functional insights into the enzymatic plasticity of the SARS-CoV-2 NiRAN domain.
Mol.Cell, 83, 2023
|
|
8FIX
 
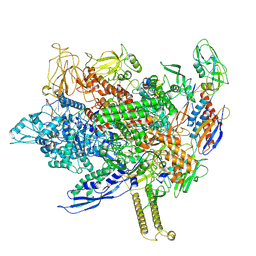 | | Cryo-EM structure of E. coli RNA polymerase backtracked elongation complex harboring a terminal mismatch | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Florez Ariza, A, Wee, L, Tong, A, Canari, C, Grob, P, Nogales, E, Bustamante, C. | | Deposit date: | 2022-12-17 | | Release date: | 2023-03-29 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | A trailing ribosome speeds up RNA polymerase at the expense of transcript fidelity via force and allostery.
Cell, 186, 2023
|
|
5E3H
 
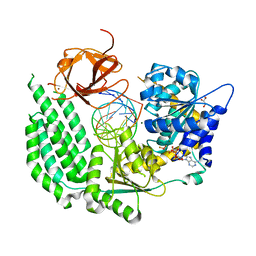 | | Structural Basis for RNA Recognition and Activation of RIG-I | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, GLYCEROL, ... | | Authors: | Jiang, F, Miller, M.T, Marcotrigiano, J. | | Deposit date: | 2015-10-02 | | Release date: | 2015-11-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of RNA recognition and activation by innate immune receptor RIG-I.
Nature, 479, 2011
|
|
3MMP
 
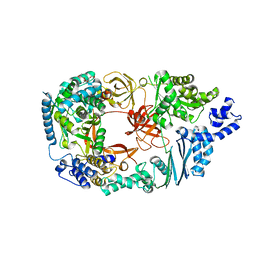 | | Structure of the Qb replicase, an RNA-dependent RNA polymerase consisting of viral and host proteins | | Descriptor: | (2S)-1-[3-{[(2R)-2-hydroxypropyl]oxy}-2,2-bis({[(2R)-2-hydroxypropyl]oxy}methyl)propoxy]propan-2-ol, Elongation factor Tu 2, Elongation factor Ts, ... | | Authors: | Kidmose, R.T, Vasiliev, N.N, Chetverin, A.B, Knudsen, C.R, Andersen, G.R. | | Deposit date: | 2010-04-20 | | Release date: | 2010-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the Qbeta replicase, an RNA-dependent RNA polymerase consisting of viral and host proteins.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
7TNY
 
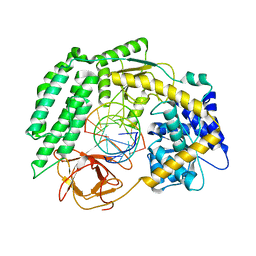 | | Cryo-EM structure of RIG-I in complex with p2dsRNA | | Descriptor: | Antiviral innate immune response receptor RIG-I, ZINC ION, p2dsRNA | | Authors: | Wang, W, Pyle, A.M. | | Deposit date: | 2022-01-22 | | Release date: | 2022-11-02 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The RIG-I receptor adopts two different conformations for distinguishing host from viral RNA ligands.
Mol.Cell, 82, 2022
|
|
7TO0
 
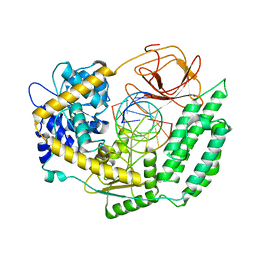 | | Cryo-EM structure of RIG-I in complex with OHdsRNA | | Descriptor: | Antiviral innate immune response receptor RIG-I, OHdsRNA, ZINC ION | | Authors: | Wang, W, Pyle, A.M. | | Deposit date: | 2022-01-22 | | Release date: | 2022-11-02 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The RIG-I receptor adopts two different conformations for distinguishing host from viral RNA ligands.
Mol.Cell, 82, 2022
|
|
7TNZ
 
 | | Cryo-EM structure of RIG-I in complex with p1dsRNA | | Descriptor: | Antiviral innate immune response receptor RIG-I, ZINC ION, p1dsRNA | | Authors: | Wang, W, Pyle, A.M. | | Deposit date: | 2022-01-22 | | Release date: | 2022-11-02 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | The RIG-I receptor adopts two different conformations for distinguishing host from viral RNA ligands.
Mol.Cell, 82, 2022
|
|
5TC1
 
 | | In situ structures of the genome and genome-delivery apparatus in ssRNA bacteriophage MS2 | | Descriptor: | Capsid protein, Maturation protein, phage MS2 genome | | Authors: | Dai, X.H, Li, Z.H, Lai, M, Shu, S, Du, Y.S, Zhou, Z.H, Sun, R. | | Deposit date: | 2016-09-13 | | Release date: | 2016-12-07 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | In situ structures of the genome and genome-delivery apparatus in a single-stranded RNA virus.
Nature, 541, 2017
|
|
