3ZDC
 
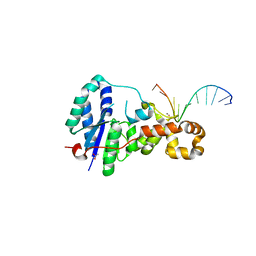 | | Structure of E. coli ExoIX in complex with the palindromic 5ov4 DNA oligonucleotide, potassium and calcium | | Descriptor: | 5OV4 DNA, 5'-D(*AP*AP*AP*AP*GP*CP*GP*TP*AP*CP*GP*CP)-3', ACETATE ION, ... | | Authors: | Hemsworth, G.R, Anstey-Gilbert, C.S, Flemming, C.S, Hodskinson, M.R.G, Zhang, J, Sedelnikova, S.E, Stillman, T.J, Sayers, J.R, Artymiuk, P.J. | | Deposit date: | 2012-11-26 | | Release date: | 2013-07-10 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | The Structure of E. Coli Exoix - Implications for DNA Binding and Catalysis in Flap Endonucleases
Nucleic Acids Res., 41, 2013
|
|
3ZD8
 
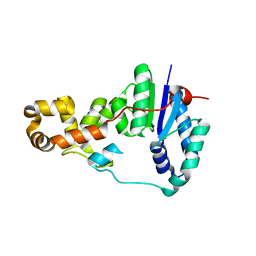 | | Potassium bound structure of E. coli ExoIX in P1 | | Descriptor: | POTASSIUM ION, PROTEIN XNI | | Authors: | Anstey-Gilbert, C.S, Hemsworth, G.R, Flemming, C.S, Hodskinson, M.R.G, Zhang, J, Sedelnikova, S.E, Stillman, T.J, Sayers, J.R, Artymiuk, P.J. | | Deposit date: | 2012-11-26 | | Release date: | 2013-07-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Structure of E. Coli Exoix - Implications for DNA Binding and Catalysis in Flap Endonucleases
Nucleic Acids Res., 41, 2013
|
|
4ALX
 
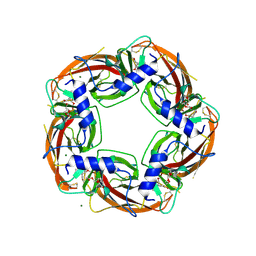 | | Crystal Structure of Ls-AChBP complexed with the potent nAChR antagonist DHbE | | Descriptor: | (4bS,6S)-6-methoxy-1,4,6,7,9,10,12,13-octahydro-3H,5H-pyrano[4',3':3,4]pyrido[2,1-i]indol-3-one, ACETYLCHOLINE BINDING PROTEIN, MAGNESIUM ION, ... | | Authors: | Shahsavar, A, Kastrup, J.S, Nielsen, E.O, Kristensen, J.L, Gajhede, M, Balle, T. | | Deposit date: | 2012-03-06 | | Release date: | 2012-08-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Lymnaea Stagnalis Achbp Complexed with the Potent Nachr Antagonist Dh-Betab-E Suggests a Unique Mode of Antagonism
Plos One, 7, 2012
|
|
4ERM
 
 | |
5NQI
 
 | |
2OS3
 
 | | Structures of actinonin bound peptide deformylases from E. faecalis and S. pyogenes | | Descriptor: | ACTINONIN, COBALT (II) ION, Peptide deformylase | | Authors: | Kim, E.E, Kim, K.-H, Moon, J.H, Choi, K, Lee, H.K, Parh, H.S. | | Deposit date: | 2007-02-05 | | Release date: | 2008-03-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structures of actinonin bound peptide deformylases from E. faecalis and S. pyogenes
To be Published
|
|
4FXE
 
 | |
1I5V
 
 | | SOLUTION STRUCTURE OF 2-(PYRIDO[1,2-E]PURIN-4-YL)AMINO-ETHANOL INTERCALATED IN THE DNA DUPLEX D(CGATCG)2 | | Descriptor: | 2-(PYRIDO[1,2-E]PURIN-4-YL)AMINO-ETHANOL, 5'-D(*CP*GP*AP*TP*CP*G)-3' | | Authors: | Favier, A, Blackledge, M, Simorre, J.P, Marion, D, Debousy, J.C. | | Deposit date: | 2001-03-01 | | Release date: | 2001-03-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of 2-(pyrido[1,2-e]purin-4-yl)amino-ethanol intercalated in the DNA duplex d(CGATCG)2.
Biochemistry, 40, 2001
|
|
4TTG
 
 | | Beta-galactosidase (E. coli) in the presence of potassium chloride. | | Descriptor: | Beta-galactosidase, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Juers, D.H. | | Deposit date: | 2014-06-20 | | Release date: | 2015-03-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Elucidating factors important for monovalent cation selectivity in enzymes: E. coli beta-galactosidase as a model.
Phys Chem Chem Phys, 17, 2015
|
|
2OS1
 
 | | Structures of actinonin bound peptide deformylases from E. faecalis and S. pyogenes | | Descriptor: | ACTINONIN, NICKEL (II) ION, Peptide deformylase, ... | | Authors: | Kim, E.E, Kim, K.-H, Moon, J.H, Choi, K, Lee, H.K, Park, H.S. | | Deposit date: | 2007-02-05 | | Release date: | 2008-03-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structures of actinonin bound peptide deformylases from E. faecalis and S. pyogenes
To be Published
|
|
2OS0
 
 | | Structures of actinonin bound peptide deformylases from E. faecalis and S. pyogenes | | Descriptor: | NICKEL (II) ION, Peptide deformylase, SULFATE ION | | Authors: | Kim, E.E, Kim, K.-H, Moon, J.H, Choi, K, Lee, H.K, Park, H.S. | | Deposit date: | 2007-02-05 | | Release date: | 2008-03-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structures of actinonin bound peptide deformylases from E. faecalis and S. pyogenes
To be Published
|
|
1NNX
 
 | | Structure of the hypothetical protein ygiW from E. coli. | | Descriptor: | Protein ygiW, SULFATE ION | | Authors: | Lehmann, C, Galkin, A, Pullalarevu, S, Sarikaya, E, Krajewski, W, Lim, K, Howard, A, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 2003-01-14 | | Release date: | 2004-03-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of the hypothetical protein ygiW from E. coli.
To be Published
|
|
4RFA
 
 | | Crystal structure of cyclic nucleotide-binding domain containing protein from Listeria monocytogenes EGD-e | | Descriptor: | Lmo0740 protein | | Authors: | Filippova, E.V, Minasov, G, Kiryukhina, O, Jedrzejczak, R, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-09-25 | | Release date: | 2014-10-15 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Crystal structure of cyclic nucleotide-binding domain containing protein from Listeria monocytogenes EGD-e
To be Published
|
|
2R69
 
 | | Crystal structure of Fab 1A1D-2 complexed with E-DIII of Dengue virus at 3.8 angstrom resolution | | Descriptor: | Heavy chain of 1A1D-2, Light chain of 1A1D-2, Major envelope protein E | | Authors: | Lok, S.M, Kostyuchenko, V.K, Nybakken, G.E, Holdaway, H.A, Battisti, A.J, Sukupolvi-petty, S, Sedlak, D, Fremont, D.H, Chipman, P.R, Roehrig, J.T, Diamond, M.S, Kuhn, R.J, Rossmann, M.G. | | Deposit date: | 2007-09-05 | | Release date: | 2007-12-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Binding of a neutralizing antibody to dengue virus alters the arrangement of surface glycoproteins.
Nat.Struct.Mol.Biol., 15, 2008
|
|
3GS7
 
 | | Human transthyretin (TTR) complexed with (E)-3-(2-methoxybenzylideneaminooxy)propanoic acid (inhibitor 13) | | Descriptor: | 3-({[(1Z)-(2-methoxyphenyl)methylidene]amino}oxy)propanoic acid, Transthyretin | | Authors: | Mohamedmohaideen, N.N, Palaninathan, S.K, Orlandini, E, Sacchettini, J.C. | | Deposit date: | 2009-03-26 | | Release date: | 2009-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Novel transthyretin amyloid fibril formation inhibitors: synthesis, biological evaluation, and X-ray structural analysis.
Plos One, 4, 2009
|
|
3ZD9
 
 | | Potassium bound structure of E. coli ExoIX in P21 | | Descriptor: | POTASSIUM ION, PROTEIN XNI | | Authors: | Anstey-Gilbert, C.S, Hemsworth, G.R, Flemming, C.S, Hodskinson, M.R.G, Zhang, J, Sedelnikova, S.E, Stillman, T.J, Sayers, J.R, Artymiuk, P.J. | | Deposit date: | 2012-11-26 | | Release date: | 2013-07-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Structure of E. Coli Exoix - Implications for DNA Binding and Catalysis in Flap Endonucleases
Nucleic Acids Res., 41, 2013
|
|
1JYS
 
 | | Crystal Structure of E. coli MTA/AdoHcy Nucleosidase | | Descriptor: | ADENINE, MTA/SAH nucleosidase | | Authors: | Lee, J.E, Cornell, K.A, Riscoe, M.K, Howell, P.L. | | Deposit date: | 2001-09-13 | | Release date: | 2002-10-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of E. coli 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase reveals similarity to the purine nucleoside phosphorylases.
Structure, 9, 2001
|
|
4KPE
 
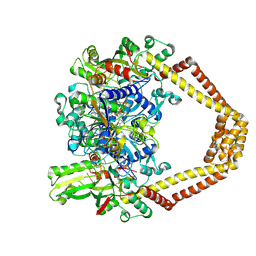 | | Novel fluoroquinolones in complex with topoisomerase IV from S. pneumoniae and E-site G-gate | | Descriptor: | (7aR,8R)-8-amino-4-cyclopropyl-12-fluoro-1-oxo-4,7,7a,8,9,10-hexahydro-1H-pyrrolo[1',2':1,7]azepino[2,3-h]quinoline-2-carboxylic acid, DNA topoisomerase 4 subunit A, DNA topoisomerase 4 subunit B, ... | | Authors: | Laponogov, I, Pan, X.-S, Vesekov, D.A, Cirz, R.T, Wagman, A.S, Moser, H.E, Fisher, L.M, Sanderson, M.R. | | Deposit date: | 2013-05-13 | | Release date: | 2014-11-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.43 Å) | | Cite: | Exploring the active site of the Streptococcus pneumoniae topoisomerase IV-DNA cleavage complex with novel 7,8-bridged fluoroquinolones.
Open Biol, 6, 2016
|
|
1YVT
 
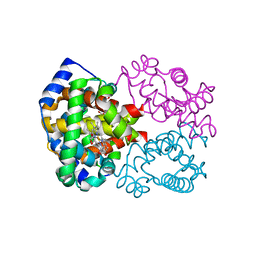 | | The high salt (phosphate) crystal structure of CO Hemoglobin E (Glu26Lys) at physiological pH (pH 7.35) | | Descriptor: | CARBON MONOXIDE, GLYCEROL, Hemoglobin alpha chain, ... | | Authors: | Malashkevich, V.N, Balazs, T.C, Almo, S.C, Hirsch, R.E. | | Deposit date: | 2005-02-16 | | Release date: | 2006-02-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The high salt (phosphate) crystal structure of CO Hemoglobin E (Glu26Lys) at physiological pH (pH 7.35)
To be Published
|
|
4KPF
 
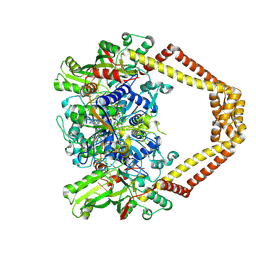 | | Novel fluoroquinolones in complex with topoisomerase IV from S. pneumoniae and E-site G-gate | | Descriptor: | (3aS,4R)-4-amino-13-cyclopropyl-8-fluoro-10-oxo-3a,4,5,6,10,13-hexahydro-1H,3H-pyrrolo[2',1':3,4][1,4]oxazepino[5,6-h]quinoline-11-carboxylic acid, E-site1, E-site2, ... | | Authors: | Laponogov, I, Pan, X.-S, Vesekov, D.A, Cirz, R.T, Wagman, A.S, Moser, H.E, Fisher, L.M, Sanderson, M.R. | | Deposit date: | 2013-05-13 | | Release date: | 2014-11-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | Exploring the active site of the Streptococcus pneumoniae topoisomerase IV-DNA cleavage complex with novel 7,8-bridged fluoroquinolones.
Open Biol, 6, 2016
|
|
1EYF
 
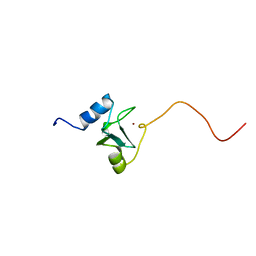 | | REFINED STRUCTURE OF THE DNA METHYL PHOSPHOTRIESTER REPAIR DOMAIN OF E. COLI ADA | | Descriptor: | ADA REGULATORY PROTEIN, ZINC ION | | Authors: | Lin, Y, Dotsch, V, Wintner, T, Peariso, K, Myers, L.C, Penner-Hahn, J.E, Verdine, G.L, Wagner, G. | | Deposit date: | 2000-05-06 | | Release date: | 2003-09-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the functional switch of the E. coli Ada protein
Biochemistry, 40, 2001
|
|
2A0F
 
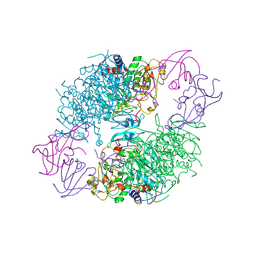 | | Structure of D236A mutant E. coli Aspartate Transcarbamoylase in presence of Phosphonoacetamide at 2.90 A resolution | | Descriptor: | Aspartate carbamoyltransferase catalytic chain, Aspartate carbamoyltransferase regulatory chain, PHOSPHONOACETAMIDE, ... | | Authors: | Stieglitz, K.A, Dusinberre, K.J, Cardia, J.P, Tsuruta, H, Kantrowitz, E.R. | | Deposit date: | 2005-06-16 | | Release date: | 2005-09-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of the E.coli Aspartate Transcarbamoylase Trapped in the Middle of the Catalytic Cycle.
J.Mol.Biol., 352, 2005
|
|
3ZDA
 
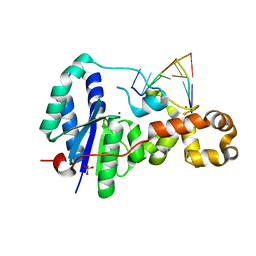 | | Structure of E. coli ExoIX in complex with a fragment of the Flap1 DNA oligonucleotide, potassium and magnesium | | Descriptor: | 5'-D(*AP*AP*GP*CP*GP*CP)-3', 5'-D(*GP*CP*GP*CP)-3', MAGNESIUM ION, ... | | Authors: | Hemsworth, G.R, Anstey-Gilbert, C.S, Flemming, C.S, Hodskinson, M.R.G, Zhang, J, Sedelnikova, S.E, Stillman, T.J, Sayers, J.R, Artymiuk, P.J. | | Deposit date: | 2012-11-26 | | Release date: | 2013-07-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Structure of E. Coli Exoix - Implications for DNA Binding and Catalysis in Flap Endonucleases
Nucleic Acids Res., 41, 2013
|
|
1TGV
 
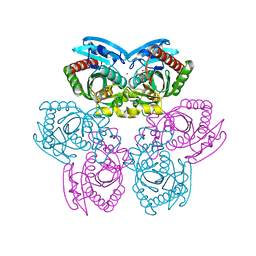 | | Structure of E. coli Uridine Phosphorylase complexed with 5-Fluorouridine and sulfate | | Descriptor: | 5-FLUOROURIDINE, POTASSIUM ION, SULFATE ION, ... | | Authors: | Bu, W, Settembre, E.C, Sanders, J.M, Begley, T.P, Ealick, S.E. | | Deposit date: | 2004-05-31 | | Release date: | 2005-06-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of E. coli Uridine Phosphorylase
To be Published, 2004
|
|
4ERP
 
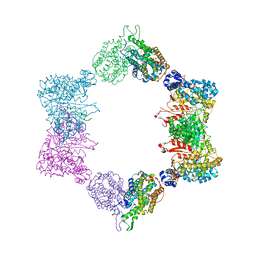 | |
